The discovery of microRNAs (miRNAs) has revealed a new level in gene expression post-transcriptional control. miRNAs are noncoding RNA molecules with 22 nucleotides that normally inhibit mRNAs by binding to their 3' untranslated region.1,2 miRNAs are translated as primordial miRNA transcripts (pri-miRNAs), which have a hairpin-like shape and are responsible for the production of mature miRNAs.3
More than 1500 human miRNAs have been found so far, as well as hundreds of viral miRNAs, each of which has the ability to target dozens of genes. Individual miRNAs have been discovered to be engaged in more than one setting, indicating that miRNAs are vital for the control of several biological activities. Overall, miRNA expression data are quite accurate in diagnosing different tumors and providing information about lineage and differentiation state. This is a common misunderstanding.4 As miR-223 affects the differentiation of numerous critical components in the innate immune response (e.g., neutrophils, monocytes, and granulocytes), it is expected that miR-223 will play a role in the early stages of infection.5 Given its profound impact on the immune system as a whole, more research on miR-223 and its role in infection is expected to be published.6–8 Excessive secretion of inflammatory components such as interferons and pro-inflammatory cytokines may result from an active immune system. As a result, immune responses are closely controlled at several levels. miRNAs were also significant regulatory factors and immune system fine tuners.8 Several studies have been published to date looking at the relationship between miRNAs and viral (swine flu, HIV, and hepatitis B) or bacterial infections.6–8 Host-directed therapy (HDT) is a relatively new strategy of infectious illness treatment that focuses on directly affecting host components and machinery that play critical roles in pathogen invasion and survival.9 Previous research has indicated miRNAs for HDT in bacterial infections, however the method of evaluating miRNAs for HDT is still in its early stages.10,11
With increased scientific output and unparalleled access to information, the need for a method of independently reviewing and analyzing research output becomes obvious and critical. As a result, a variety of data analysis tools and internet-based search engines have been developed to facilitate the processing and organization of this scientific output into a comprehensible form. One such method used for this assessment is bibliometric parameters. In recent years, many new bibliometric analysis approaches have begun to enter the medical literature. This method is an analytical search study and it can be extended by methods such as mapping and graphing. Many methodologies may be used to conduct these investigations, including content analysis, comparisons of scientific production by years, nations, and citation numbers. This approach, however, may be used to study other sources, such as any database, theses, journals, congresses, and so on.12–20 The major purpose of this study was to investigate the significance of publications and identify research developments and clusters using bibliometric methods.
Methods
We conducted a bibliometric and network visualization analysis study. Data was obtained from Thomson Reuters’ Web of ScienceTM (WoS) (Clarivate Analytics, Philadelphia, PA, USA) database (http://apps.webofknowledge.com/). The titles, document types, publication years, authors, affiliations, keywords, publishing journals, abstracts of each document, and citations within the WoS database were saved as TXT files and retrieved into Microsoft Office Excel 2019 (Los Angeles, CA, USA).
The Hirsch (H)-index was presented as an alternative to conventional bibliometric indicators as the best assessment for evaluating the impact of scientific research. Data for this investigation were obtained from WoS database on 10 April 2022.
We searched for titles including the terms microRNAs or miRNA and sepsis or infection or viral infection or bacterial infection published between 1970 and 31 March 2022. All peer-reviewed publications, including reviews, letters to the editor, and editorials, were accepted in all languages. Web of Science Core Collection search results included entries from the WoS Core Collection, which included the Science Citation Index Expanded (SCI-EXPANDED), the Social Sciences Citation Index (SSCI), the Arts and Humanities Citation Index (A & HCI), the Book Citation Index (BKCI), the Conference Proceedings Citation Index (CPCI), Current Chemical Reactions (CCR Expanded), and Index Chemicus (IC).
We used the WoS database’s Results Analysis and Citation Report to analyze the number of publications in various viewpoints, such as years, journals, and authors. To visualize country collaboration networks and keywords, we used the VOSviewer software version 1.6.18 for Microsoft Windows (Leiden University, Leiden, Netherlands). We created co-occurrence networks from the obtained publications’ bibliographic metadata (e.g., affiliations, citations, and keywords).
The study complied with the 2013 Helsinki Declaration. The study did not require ethics committee approval.
Results
Based on the search method utilized in this study, the findings revealed that there were 623 documents of which 251 (40.3%) were articles. One hundred percent of the documents were published in English. The published documents and citation numbers were given in Figure 1. The first document was published in 2007, and the maximum number of documents was published in 2021. The number of documents has been increasing since 2019.
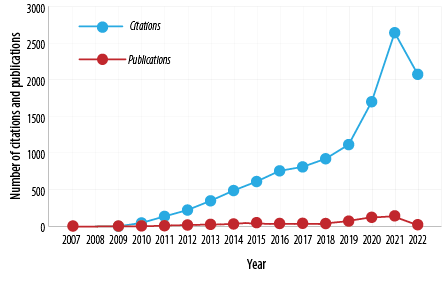 Figure 1: The number of publications and citations over the years.
Figure 1: The number of publications and citations over the years.
The largest number of studies (23.8%) were from the immunology area [Table 1].
Table 1: The number of publications on microRNAs according to research areas (N = 623).
|
Immunology
|
148
|
23.8
|
|
Cell biology
|
98
|
15.7
|
|
Research experimental medicine
|
95
|
15.2
|
|
Biochemistry molecular biology
|
80
|
12.8
|
|
Virology
|
52
|
8.3
|
|
Microbiology
|
48
|
7.7
|
|
Pharmacology pharmacy
|
44
|
7.1
|
|
Gastroenterology hepatology
|
35
|
5.6
|
|
Oncology
|
35
|
5.6
|
|
Science technology other topics
|
34
|
5.5
|
|
Biotechnology applied microbiology
|
31
|
5.0
|
|
General internal medicine
|
27
|
4.3
|
|
Infectious diseases
|
24
|
3.89
|
|
Veterinary sciences
|
22
|
3.5
|
|
Parasitology
|
21
|
3.4
|
|
Fisheries
|
20
|
3.2
|
|
Cardiovascular system cardiology
|
19
|
3.0
|
|
Surgery
|
17
|
2.7
|
|
Respiratory system
|
16
|
2.6
|
|
Marine freshwater biology
|
15
|
2.4
|
|
Pathology
|
15
|
2.4
|
|
Hematology
|
14
|
2.2
|
|
Biophysics
|
13
|
2.1
|
|
Chemistry
|
11
|
1.8
|
*Showing 25 out of 49 entries.
A total of 619 (99.4%) of the publications were published in journals indexed by SCI-EXPANDED. The documents were published in 257 different journals. The list of the top 25 journals is presented in Table 2.
Table 2: The top 25 published journals on microRNAs in infectious diseases (N = 623).
|
Inflammation
|
14
|
2.2
|
|
Journal of Immunology
|
14
|
2.2
|
|
Plos One
|
14
|
2.2
|
|
Scientific Reports
|
14
|
2.2
|
|
Experimental and Therapeutic Medicine
|
13
|
2.1
|
|
European Review for Medical and Pharmacological Sciences
|
12
|
1.9
|
|
Fish Shellfish Immunology
|
11
|
1.8
|
|
Journal of Virology
|
11
|
1.8
|
|
Molecular Medicine Reports
|
11
|
1.8
|
|
Plos Pathogens
|
10
|
1.6
|
|
Frontiers in Immunology
|
9
|
1.4
|
|
Mediators of Inflammatıon
|
9
|
1.4
|
|
Biochemical and Biophysical Research Communications
|
8
|
1.3
|
|
International Immunopharmacology
|
8
|
1.3
|
|
Shock
|
8
|
1.3
|
|
American Journal of Translational Research
|
7
|
1.1
|
|
Bioengineered
|
7
|
1.1
|
|
Biomed Research International
|
7
|
1.1
|
|
Developmental and Comparative Immunology
|
7
|
1.1
|
|
European Journal of Immunology
|
7
|
1.1
|
|
Frontiers in Cellular and Infection Microbiology
|
7
|
1.1
|
|
Hepatology
|
7
|
1.1
|
|
Journal of Cellular and Molecular Medicine
|
7
|
1.1
|
|
Journal of Hepatology
|
7
|
1.1
|
*Showing 25 out of 257 entries.
China dominated scientific production on miRNAs in infectious diseases with 398 (63.9%) publications. The top five leading scientifically productive countries on this topic also included the USA (n = 100; 16.1%), Japan (n = 24; 3.9%), Germany (n = 20; 3.2%), and Italy (n = 17; 2.7%). The publications were from 43 countries [Table 3].
Table 3: The list of most published countries on microRNAs (N = 623).
|
China
|
398
|
63.9
|
|
USA
|
100
|
16.1
|
|
Japan
|
24
|
3.9
|
|
Germany
|
20
|
3.2
|
|
Italy
|
17
|
2.7
|
|
India
|
15
|
2.4
|
|
Egypt
|
13
|
2.1
|
|
England
|
13
|
2.1
|
|
Australia
|
12
|
1.9
|
|
Brazil
|
10
|
1.6
|
|
Canada
|
7
|
1.1
|
|
France
|
7
|
1.1
|
|
Iran
|
7
|
1.1
|
|
Thailand
|
7
|
1.1
|
|
Taiwan
|
6
|
1.0
|
|
Netherlands
|
5
|
0.8
|
|
South Korea
|
5
|
0.8
|
|
Spain
|
5
|
0.8
|
|
Denmark
|
4
|
0.6
|
|
Swıtzerland
|
4
|
0.6
|
|
Mexico
|
3
|
0.5
|
|
Sweden
|
3
|
0.5
|
|
Belgium
|
2
|
0.3
|
|
Chile
|
2
|
0.3
|
*Showing 25 out of 43 entries; 13 records (2.1%) do not contain data in the field being analyzed.
The Nanjing Medical University from China was the leading affiliation on this topic with 24 documents [Table 4].
Table 4: The list of universities with more than 10 publications on microRNAs in infectious diseases research area (N = 623).
|
Nanjing Medical University
|
24
|
3.9
|
|
The Chinese Academy of Sciences
|
17
|
2.7
|
|
Sun Yat Sen University
|
16
|
2.6
|
|
Wuhan University
|
14
|
2.2
|
|
League of European Research Universities, Leru
|
13
|
2.1
|
|
Zhejiang University
|
13
|
2.1
|
|
Zhengzhou University
|
13
|
2.1
|
|
Egyptian Knowledge Bank
|
12
|
1.9
|
|
Shanghai Jiao Tong University
|
11
|
1.8
|
|
East Tennessee State University
|
10
|
1.6
|
The National Natural Science Foundation of China (n = 145, 23.3%), the National Institutes of Health (n = 51; 8.2%), and the United States Department of Health and Human Services (n = 51; 8.2%) were the top funding sponsors.
The documents were cited 10 370 times in total (16.7 per document). The H-index was 50. The documents originated from China were cited 5705 times (average: 14.3/document). The documents originated from the USA were cited 2190 times (average: 21.9/document). The top cited 10 publications were summarized in Table 5.
Table 5: Top 10 cited publications on microRNAs in infectious diseases.
|
1
|
478
|
The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma
|
Ma et al.
|
Nature Immunology
|
2011
|
25.606
|
|
2
|
255
|
miR-223: infection, inflammation and cancer
|
Haneklaus et al.
|
Journal of Internal Medicine
|
2013
|
8.989
|
|
3
|
252
|
Serum miR-146a and miR-223 as potential new biomarkers for sepsis
|
Wang et al.
|
Biochemical and Biophysical Research Communications
|
2010
|
3.575
|
|
4
|
238
|
MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis
|
Vasilescu et al.
|
Plos One
|
2009
|
3.24
|
|
5
|
213
|
Exosomal miR-146a contributes to the enhanced therapeutic efficacy of interleukin-1-primed mesenchymal stem cells against sepsis
|
Song et al.
|
Stem Cells
|
2017
|
6.277
|
|
6
|
200
|
Small molecule modifiers of miR-122 function for the treatment of HCV infection and hepatocellular carcinoma
|
Young et al.
|
Journal of the American Chemical Society
|
2010
|
15.419
|
|
7
|
187
|
Exosomal miR-223 contributes to mesenchymal stem cell-elicited cardioprotection in polymicrobial sepsis
|
Wang et al.
|
Scientific Reports
|
2015
|
4.38
|
|
8
|
174
|
Oncogenic HPV infection interrupts the expression of tumor-suppressive miR-34a through viral oncoprotein E6
|
Wang et al.
|
RNA
|
2009
|
4.942
|
|
9
|
145
|
Serum miR-122 as a biomarker of necroinflammation in patients with chronic HCV infection
|
Bihrer et al.
|
The American Journal of Gastroenterology
|
2011
|
10.864
|
A visualization analysis of co-authorship among mostly published authors is given in Figure 2. The cooperation between the most published authors is indicated by the lines connecting the circles, the wider the lines, the closer the cooperation.
Several authors engaged in active collaboration. However, the majority of authors are dispersed and lack consistent, close-knit contact and cooperative relationships.
The map of keywords co-occurrence was generated with VosViewer in Figure 3. According to the frequency and centrality, we found the most popular keyword was ῾sepsis’. The circles connecting lines represent the cooperation between the most popular keywords, the wider the lines, the tighter the cooperation.
 Figure 2: Visualization analysis of co-authorship among most publishing authors.
Figure 2: Visualization analysis of co-authorship among most publishing authors.
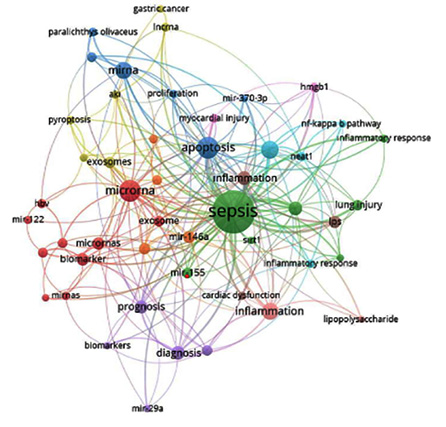 Fıgure 3: Keywords analysis.
Fıgure 3: Keywords analysis.
Figure 4 depicts the bibliographic coupling of universities. Each circle in the drawing symbolizes an institution, and its size corresponds to the volume of publications it produces. The lines separating the circles represent university collaboration, the larger the lines, the closer the cooperation.
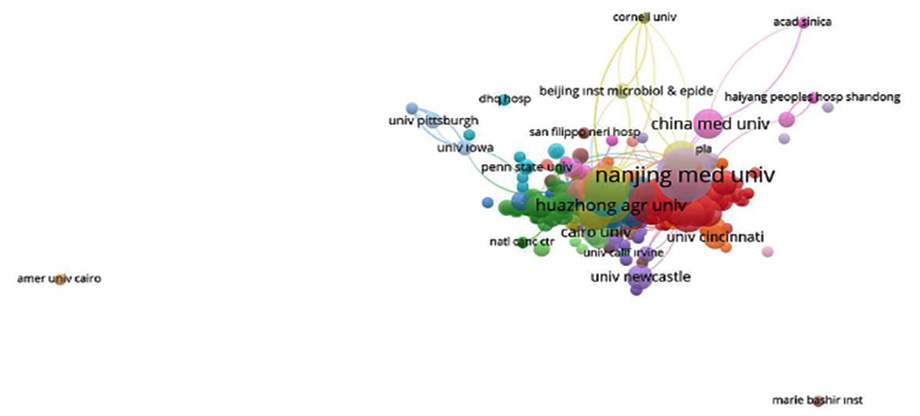 Figure 4: The bibliographic coupling of universities.
Figure 4: The bibliographic coupling of universities.
Discussion
The discovery of deregulated miRNA expression raises the possibility that miRNAs may be employed as diagnostic or prognostic indicators of a variety of diseases. Furthermore, miRNAs are appealing therapeutic targets for a variety of diseases, including cancer. The new role of miRNAs and their involvement in a variety of activities has sparked a surge in scientific interest in miRNA function. Using bibliometrics, the field of miRNA study provides an amazing chance to trace the evolution of a novel field of research inquiry from the point of discovery to a quickly developing field.20–24 However, there are a limited number of bibliometric studies on miRNAs.20–25 No similar bibliometric study on miRNAs in the infectious diseases research area was available in the literature. To the best of our knowledge, this is the first bibliometric analysis of the miRNA area. The first bibliometric study on miRNAs was published in 2015.20 Our study showed that annual publications have a growing trend in research output, especially since 2019.
Databases that allow simple and extensive data analysis, such as Pubmed, EBSCO, Scopus, Pro-Quest, and the WoS databases, are often used for bibliometric studies.12–20 We used the WoS database, which is one of the most prestigious databases. Many different tools can be performed for visualization. In a similar study,24 CiteSpace was used, but we preferred VOSviewer.
The miRNA was first isolated in 1993 from the nematode Caenorhabditis elegans.26 The term ῾miRNA’ was coined by Ruvkun in 2001 to denote a class of naturally occurring short, non-coding RNA molecules 19 to 21 nucleotides long.27 According to our findings, the first document was published in 2007. In our research, 40.3% of all documents are original articles. This may be due to the scientific interest in this new topic.
While 43 countries have contributed to miRNA in infection literature to date, five countries dominate scientific production in this research field. China dominated scientific production on miRNAs in infection with 398 (63.9%) publications. The remaining top five leading scientifically productive countries on this topic were the USA, Japan, Germany, and Italy. Documents originating from China were cited 5705 times (average: 14.3/document). Documents originating from the USA were cited 2190 times (average: 21.9/document). This may mean that the scientific quality of publications originating from the USA is higher. An interesting finding of our study is that China is the leading country in this regard, and the importance given by Chinese funders and universities to this issue has been revealed. While the USA was in first place in various previous medical bibliometric studies,12–15,17 China took the first place in bibliometric studies on miRNA.20–24
Special events in the miRNA field can be found by analyzing the most cited documents since the discovery of this study topic. Our study reviewed the 10 most cited primary studies that demonstrate the ongoing main findings in the field of miRNA infection. Two of the 10 most cited documents were published in the Nature Immunology journal. The top 10 most cited primary documents had an average journal impact factor of 42.0, demonstrating their exposure and importance as a driver of exponential growth in annual publications.
Cooperative network analysis reveals that the productive authors and institutions in this discipline each have a large number of collaborators, but they do not collaborate directly with one another. This might be due to their capacity to perform solid independent research. They may already possess the qualities of a great partner that others want, obviating the need to actively seek additional collaboration.24 The mapping analyses of our study and collaborations were defined in Figures 2 and 4.
The literature network is made up of several clusters, and VOSviewer labels each one by picking representative terms from the cited literature. We can establish the research hotspots and trends of a certain era by evaluating the citation network and keywords in the temporal dimension to determine the rise, flourishing, and fall of specific research clusters.24 The more publications in a cluster in the timeline [Figure 3], the more important the cluster is and the larger its position in the figure. The research hotspots of miRNAs in infectious diseases have mostly concentrated on the following elements during the last decade, based on an examination of the keywords of each cluster and the top articles in each cluster. The diagnosis, severity, and etiology of sepsis have all been investigated using a variety of clinical and serum-based markers. Few of these factors could, however, be applied clinically up until this point. Instead of encoding proteins, miRNAs control gene expression by preventing the translation or transcription of the mRNAs they target. Recent research has shown that miRNAs are released into the bloodstream and that different pathologic conditions like inflammation, infection, and sepsis may alter the range of circulating miRNAs.6,8
VOSviewer was used in this instance to create the map of keyword co-occurrence. The most common keyword, as determined by frequency and centrality, was ῾sepsis’. Using bibliometric techniques, we discovered several deregulated miRNAs, such as miR-122, miR-133a, miR-146, miR-155, and miR-370, which were discussed in the context of sepsis and infection. However, due to the widespread use of synonym keywords in publications, we were obliged to employ keyword groupings rather than each individual keyword. The journal might want to adopt a more uniform method for author keyword selection. To do this, the publication might offer authors a drop-down list of keywords to choose from in the submission process. This strategy might have advantages that go beyond making bibliometric analysis easier. Standardizing terms, for instance, would make it easier for readers to locate items they are interested in.
This is the first bibliometric analysis to examine the trends of miRNAs in the field of infectious diseases. Furthermore, there are several limitations to this bibliometric analysis. The electronic database is confined to the WoS database, and other electronic databases, such as PubMed, Embase, Scopus, and the Cochrane Library, were not searched and evaluated. Another drawback is that the data for 2022 is insufficient because the year has not yet ended.
Conclusion
Since 2019, the number of studies on miRNA in infectious diseases has gradually increased. China and the USA have made tremendous contributions to work in this area. Understanding these important factors, as well as how research is conducted and guided, could lead to a new perspective in formulating new strategies to manage various infections in the years to come.
Disclosure
The authors declared no conflicts of interest. No funding was received for this study.
references
- 1. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004 Jan;116(2):281-297.
- 2. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 2008 Feb;9(2):102-114.
- 3. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010 Sep;11(9):597-610.
- 4. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature 2005 Jun;435(7043):834-838.
- 5. Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008 Feb;451(7182):1125-1129.
- 6. Haneklaus M, Gerlic M, O’Neill LA, Masters SL. miR-223: infection, inflammation and cancer. J Intern Med 2013 Sep;274(3):215-226.
- 7. Loureiro D, Tout I, Narguet S, Benazzouz SM, Mansouri A, Asselah T. miRNAs as potential biomarkers for viral hepatitis B and C. Viruses 2020 Dec;12(12):1440.
- 8. Mishra R, Krishnamoorthy P, Kumar H. MicroRNA-30e-5p regulates SOCS1 and SOCS3 during bacterial infection. Front Cell Infect Microbiol 2021 Jan;10:604016.
- 9. Kaufmann SH, Dorhoi A, Hotchkiss RS, Bartenschlager R. Host-directed therapies for bacterial and viral infections. Nat Rev Drug Discov 2018 Jan;17(1):35-56.
- 10. Iannaccone M, Dorhoi A, Kaufmann SH. Host-directed therapy of tuberculosis: what is in it for microRNA? Expert Opin Ther Targets 2014 May;18(5):491-494.
- 11. Sabir N, Hussain T, Shah SZ, Peramo A, Zhao D, Zhou X. miRNAs in tuberculosis: new avenues for diagnosis and host-directed therapy. Front Microbiol 2018 Mar;9:602.
- 12. Can ÖZ. Scopus veri tabanına dayalı bibliyometrik değerlendirme: miyelodisplastik sendrom konulu yayınların global analizi ve türkiye kaynaklı yayınların değerlendirilmesi. Journal of Biotechnology and Strategic Health Research 2021;5(2):125-131.
- 13. Küçük U, Alkan S, Uyar C. Bibliometric analysis of infective endocarditis. Iberoam J Med 2021;3(4):350-355.
- 14. García-Gómez F, Ramírez-Méndez F. Análisis bibliométrico de la Revista Médica del IMSS en la base de datos Scopus durante el periodo 2005-2013. Rev Med Inst Mex Seguro Soc 2015 May-Jun;53(3):323-335.
- 15. Sweileh WM. Bibliometric analysis of peer-reviewed literature on climate change and human health with an emphasis on infectious diseases. Global Health 2020 May;16(1):44.
- 16. Yıldız E. Türkiye’de gebelik ve anestezi konulu yayınların bibliyometrik analizi. Black Sea Journal of Health Science 2022;5(1):50-55.
- 17. Köylüoğlu AN, Aydın B, Özlü C. Bibliometric evaluation based on Scopus database: global analysis of publications on diabetic retinopathy and comparison with publications from Turkey. D J Med Sci 2021;7(3):268-275.
- 18. Durgun C, Alkan S, Durgun M, Demiray EK. Türkiye’den kist hidatik konusunda yapılmış yayınların analizi. Black Sea Journal of Health Science 2022;5(1):45-49.
- 19. Aysun Ö. Bibliometric analysis of publications on pulmonary rehabilitation. Black Sea Journal of Health Science 2022;5(2):7-8.
- 20. Casey MC, Kerin MJ, Brown JA, Sweeney KJ. Evolution of a research field-a micro (RNA) example. PeerJ 2015 Mar;3:e829.
- 21. Yun L, Wang L, Pan Y, Liu M, Han Q, Sun J, et al. Current status and development trend of miRNAs in osteoporosis-related research: a bibliometric analysis. Folia Histochem Cytobiol 2021;59(4):203-211.
- 22. Shaw P, Lokhotiya K, Kumarasamy C, Sunil K, Suresh D, Shetty S, et al. Mapping research on miRNAs in cancer: a global data analysis and bibliometric profiling analysis. Pathophysiology 2022 Feb;29(1):66-80.
- 23. Manoel Alves J, Handerson Gomes Teles R, do Valle Gomes Gatto C, Muñoz VR, Regina Cominetti M, Garcia de Oliveira Duarte AC. Mapping research in the obesity, adipose tissue, and microRNA field: a bibliometric analysis. Cells 2019 Dec;8(12):1581.
- 24. Gao K, Dou Y, Lv M, Zhu Y, Hu S, Ma P. Research hotspots and trends of microRNA in periodontology and dental implantology: a bibliometric analysis. Ann Transl Med 2021 Jul;9(14):1122.
- 25. Hu WS, Zhang Q, Li SH, Ai SC, Wu QF. Ten hotspot microRNAs and their potential targets of chondrocytes were revealed in osteoarthritis based on bibliometric analysis. J Healthc Eng 2022 Apr;2022:8229148.
- 26. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993 Dec;75(5):843-854.
- 27. Ruvkun G. Molecular biology. Glimpses of a tiny RNA world. Science 2001 Oct;294(5543):797-799.