Stenotrophomonas maltophilia is a gram-negative, non-fermentative, catalase-positive, and oxidase-negative bacterium.1 It is an opportunistic nosocomial pathogen and infections associated with it are difficult to control, being multiple drug resistant.2 This bacterium is widely distributed in nature and has been isolated from humans, animals, and hospital environment.3,4 S. maltophilia is a common cause of infections in patients with cystic fibrosis, cancer, neutropenia, intravenous catheterization, and patients with a history of multiple antibiotic use.5
S. maltophilia has also been recognized as a cause of nosocomial bacteremia in intensive care units (ICUs) and in immunocompromised patients. This bacterium can cause 20–75% of deaths in the case of pneumonia and 20–60% of cases of bacteremia.6–8 Children and infants are often susceptible to S. maltophilia infection. Many factors can contribute to infections, especially bloodstream infections (BSI) in hospitalized children. There are few epidemiological studies on S. maltophilia infections in Iranian children.
Treatment options for S. maltophilia infection are limited due to the pathogen’s innate resistance to most antibiotics. Trimethoprim-sulfamethoxazole combination is the current therapy of choice.9,10 Biofilm formation is known to be a preferred survival strategy for S. maltophilia, in addition to tolerance to high doses of antibiotics. Through biofilm production, S. maltophilia strains can readily adhere to the surfaces in hospitals, facilitating transmission.11
Nowadays, molecular typing is widely used to study the transmission routes of bacterial infections, especially nosocomial infections. Polymerase chain reaction (PCR)-based molecular typing method has advantages such as high speed, simplicity, and low cost. Among the PCR-based molecular typing methods, repetitive extragenic palindromic (rep)-PCR is often preferred due to its low cost and rapidity.12
Given that S. maltophilia bacteremia is an emerging infection of concern associated with high mortality in immunocompromised hospitalized pediatric patients, this study aimed to evaluate the frequency, antibiotic resistance patterns, biofilm- formation ability, and prevalence of biofilm-related genes, as well as the genetic relationships of S. maltophilia strains isolated from blood cultures of hospitalized children in Iran.
Methods
In this cross-sectional study, 450 oxidase-negative bacilli were isolated from the blood cultures of pediatric inpatients hospitalized in two western Iranian cities of Hamadan and Borujerd from June 2020 to June 2021. Identification of S. maltophilia was made using standard microbiological tests and a biochemical identification kit (Microgen GN-B kit). S. maltophilia isolates were also confirmed by PCR using 16S rRNA primer.13 This study was approved by the ethics committee of Hamadan University of Medical Sciences (Ref. IR.UMSHA. REC. 1399.092).
Antibiotic susceptibility of S. maltophilia strains was determined by the Kirby–Bauer disk diffusion method and Etest. The antibiotic panel included trimethoprim/sulfamethoxazole (TMP/SMX: 1.25/23.75 µg), levofloxacin (LEV: 5 µg), and ceftazidime (CAZ: 30 µg). Quality control was maintained using Escherichia coli ATCC 25922. Results were interpreted as stipulated by the Clinical and Laboratory Standards Institute (CLSI 2021).14
The biofilm formation of S. maltophilia isolates was investigated by microtiter plate (96-well plate) using the dye crystal violet dye. S. maltophilia biofilm quantitation was performed by a spectrophotometric method as previously described.15 All experiments were performed in triplicate.
The genomic DNA of S. maltophilia was extracted by boiling method. DNA was extracted after treating cells (colonies) with alkali (NaOH).16 The presence of S. maltophilia biofilm-related genes (rpfF, spgM, and rmlA) was detected by PCR with specific primers previously described.17
The genetic relationships of the isolates of S. maltophilia were investigated by rep-PCR typing. The rep-PCR analysis was performed with a single primer BOX-A1R (5'-CTA CGG CAA GGC GAC GCT GAC G-3'). The PCR reaction mixture consisted of a total volume of 25 μL. Thermal cycling was performed according to the following procedure: initial denaturation (94 °C for 10 min), followed by 25 cycles of denaturation (at 94 °C for 45 sec), annealing (at 50 °C for 1.5 min), extension (at 65 °C for 8 min) and a final cycle of extension at 65 °C for 16 min.18 The rep-PCR products were loaded on a 2% agarose gel at 70 V for one hour, and the repetitive sequence band patterns (REP profiles) were visualized in a gel documentation system.17 The REP patterns were analyzed by an online data analyzer accessed from insilico.ehu.eus/dice_upgma. The REP profiles were compared using the Dice method and clustered according to the unweighted paired group method with arithmetic mean (UPGMA).18
Results
In total, out of 450 oxidase-negative bacilli, 72 (16.0%) strains of S. maltophilia were identified; 30 strains (41.7%) were from hospitals in Hamadan and 42 strains (58.3%) were from hospitals in Borujerd. The ages of the patients ranged from < 1 year to 12 years. More than 80% of patients were under 10 years of age. There was a significant difference between the ages of patients from Hamadan and Borujerd hospitals (p = 0.010). The majority were male (43; 59.7%). There was no statistically significant relationship between the patients’ sex and isolates of S. maltophilia (p = 0.808).
According to the phenotypic biofilm formation assay, among the 72 strains of S. maltophilia, 19 (26.4%), 38 (52.8%), and 10 (13.9%) isolates produced strong, moderate, and weak biofilm, respectively. Five isolates (6.9%) did not produce biofilm. The frequencies of biofilm-related genes by PCR were given as follows: rmlA (59; 81.9%), rpfF (54; 75.0%), and spgM (72; 100%) [Box 1].
|
Biofilm associated genes
|
spgM
|
rmlA
|
rpfF
|
|
100
|
81.9
|
75.0
|
|
Biofilm-formation ability
|
Weak
|
Moderate
|
Strong
|
|
13.9
|
52.8
|
26.4
|
|
Antibiotic sensitivity
|
LEV
|
TMP/SMX
|
CAZ
|
LEV: levofloxacin; TMP/SMX: trimethoprim-sulfamethoxazole; CAZ: ceftazidime.
Box 1: Frequency of biofilm-related genes, biofilm-forming ability, and antibiotic susceptibility of isolates of S. maltophilia.
Antimicrobial susceptibility analysis showed that 93.1% of the S. maltophilia strains were sensitive to TPM/SMX (6.9% were intermediate). All isolates were sensitive to LEV and resistant to CAZ [Box 1].
According to the results of the rep-PCR analysis, the size of the amplicons varied from 300 bp to > 1 kb. Through analysis of the results, the genetic diversity among S. maltophilia strains was observed [Figure 1]. The rep-PCR analysis revealed 14 different REP types, which were divided into 11 common types (CTs) and three single types. CTs included 2–21 isolates [Figure 2]. Hamadan city isolates are represented by H and Borujerd city isolates are represented by B in Figure 2. The REP profiles of Hamadan and Borujerd isolates are completely different. The largest CT belonged to Hamadan containing 21 isolates [Figures 1 and 2]. Different types of REP show the same antibiotic resistance pattern. The difference between isolates is mainly related to the differences in biofilm formation strength.
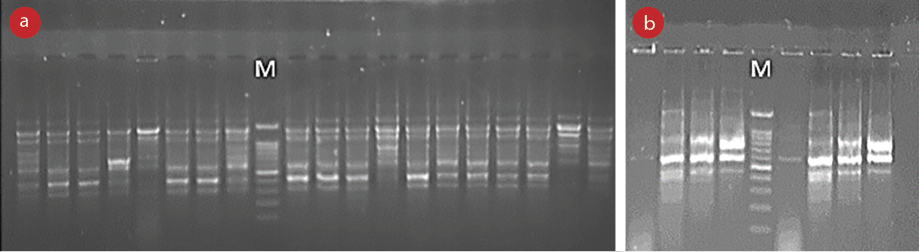 M: Marker 100 bp; REP: repetitive sequence.
M: Marker 100 bp; REP: repetitive sequence.
Figure 1: Band patterns resulting from amplification of REP regions in S. maltophilia isolates. (a) Band patterns of Hamadan city isolates and (b) band patterns of Borujerd city isolates.
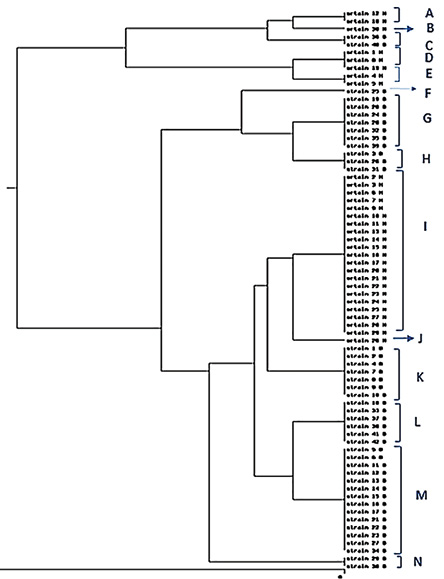 UPGMA: unweighted pair group method with arithmetic mean; rep-PCR: repetitive element sequence-based polymerase chain reaction.
UPGMA: unweighted pair group method with arithmetic mean; rep-PCR: repetitive element sequence-based polymerase chain reaction.
Figure 2: Dendrogram of rep-PCR fingerprinting of S. maltophilia isolates, comparison by Dice method and clustering by UPGMA method.
Discussion
In this study, the prevalence of S. maltophilia isolated from blood cultures of pediatric patients was 16.0%. Although this may be low compared with other hospital-acquired bacteria, it is significant due to this pathogen’s intrinsic resistance to common antimicrobial agents, presence of virulence genes, and biofilm-formation ability. There are various reports on the prevalence of S. maltophilia in Iran. In a previous study from Hamadan, 12 (4.8%) S. maltophilia were isolated from blood cultures.19 The isolates were verified by standard biochemical methods. Based on the results of that study and ours, the incidence of S. maltophilia in Hamadan hospitals has increased over time. In another study performed by Bostanghadiri et al,17 from Iran, 164 clinical isolates of S. maltophilia were identified and confirmed using standard biochemical tests and PCR. Most (83.5%) samples positive for S. maltophilia were blood cultures. As per our results, the prevalence of S. maltophilia was higher in males than in females in the ratio of 1.15 to 1.17 In another new study in Iran, 117 strains of S. maltophilia were isolated from different clinical sources. S. maltophilia isolates were identified by routine microbiological and biochemical tests. The highest S. maltophilia prevalence was observed in the blood (92.3%) and the lowest in wounds (0.85%).20 In a study by Duan et al,21 from Shanghai, China, a total of 104 strains of S. maltophilia were collected from different pediatric wards. Contrary to our results, most strains of S. maltophilia were isolated from sputum sources.21 In a retrospective cohort study of hospitalized pediatric patients in Saudi Arabia, most (88.2%) bacteremia cases were catheter-related BSI.6
In our study, over 80% of children with S. maltophilia were < 9 years old. According to several studies, hospitalized infants and children of any age can be susceptible to S. maltophilia infection.6,17,21 Various factors can contribute to infection in hospitalized children, especially BSI. Risk factors for S. maltophilia BSI may be length of ICU stay, use of mechanical ventilators, indwelling catheters, and length of hospital stay.22
TMP/SMX is considered the most effective antibiotic to treat S. maltophilia infections. However, a number of recent reports of resistance from S. maltophilia-induced infections have raised concerns over its continued efficacy. Alternative antibiotics, such as LEV and minocycline, have been reported effective against invasive S. maltophilia infections, especially in severe infections.17,23 In this study, LEV and TMP/SMX were found to be effective for S. maltophilia and CAZ as unsuitable. However, the efficacy of minocycline was not studied, because it is unavailable in Iranian hospitals and is generally not prescribed here.
Different results of antimicrobial susceptibility from ours were reported from Iran and elsewhere. In a previous study in Hamadan hospitals, all strains of S. maltophilia were found susceptible to ofloxacin (a fluoroquinolone antibiotic) and TMP/SMX; however, another study that used data from hospitals in various regions in Iran found that 91.0%, 99.3%, and 63.5% of S. maltophilia isolates were susceptible to TMP/SMX, LEV, and CAZ, respectively.17,19 In a cross-sectional study in Southwest Iran, all of the 44 S. maltophilia isolates from different clinical specimens were susceptible to TMP/SMX.24 Inconsistent with our results, a study in Tehran hospitals, a total of 150 S. maltophilia isolates were collected from various clinical specimens including respiratory specimens, secretions from ventilator-associated pneumonia, as well as from surgical instruments and catheters. Eighty percent of the isolates were resistant to TMP/SMX while 20% were resistant to a fluoroquinolone such as ofloxacin. One of the main reasons for this difference in the results compared to ours might be the differences in type of samples examined and the location of their study. Studies outside Iran also show variations resistance to TMP/SMX across geographical areas.24 However, S. maltophilia resistance to TMP/SMX has not been reported > 10% except in respiratory infections and in patients with cystic fibrosis.17,25–28
The rate of resistance to CAZ in our patients was higher than the rates previously reported from different parts of Iran and from other countries.6,17,19,27,29 This antibiotic should be considered inappropriate against S. maltophilia in children in the studied hospitals.
Another factor investigated in this study was the biofilm-forming ability of S. maltophilia strains. Biofilm formation on hospital surfaces and in human tissues is an important survival feature of S. maltophilia. In this study, most S. maltophilia isolates were biofilm producers. All isolates of S. maltophilia carried the spgM gene. However, other related-biofilm genes, rmlA and rpfF were found in 81.9% and 75.0% of the isolates, respectively. Our results are in agreement with the results of Bostanghadiri et al,17 where most of the isolates were biofilm producers, and 88.4%, 83.5%, and 100% of the isolates were positive for rmlA, rpfF, and spgM genes. In a study by Flores-Treviño et al,30 the rate of biofilm formation and isolation of potent biofilm procedure was higher than both in our study and that by Bostanghadiri et al.17 Studies have shown that the spgM gene plays an important role in the formation of strong biofilm.17,29–31
In this study, the genetic diversity of the isolates of S. maltophilia was determined inexpensively and quickly using the rep-PCR technique using a single primer,12 showing clonal diversity among S. maltophilia isolates. Our 72 isolates of S. maltophilia were classifiable into 14 different REP types. Genetic diversity in S. maltophilia isolates has been confirmed by many studies.17,20,32,33 In a study by Bostanghadiri et al,17 high clonal diversity (16 CTs and 114 single types) was detected by rep-PCR assay in S. maltophilia isolates. The reasons for their findings have a greater genetic diversity than us due to their larger sample size and the fact that their samples were sourced from different regions of Iran. Duan et al,21 identified 104 highly diverse isolates of S. maltophilia from a children’s hospital in China. They used two different molecular typing methods, pulsed-field gel electrophoresis and Multilocus sequence typing of S. maltophilia isolates, which were found divided into 93 clusters and 59 sequence types.21
There are limitations to our study. It was conducted during the COVID-19 pandemic. Therefore, we encountered challenges in taking samples. Clinical information of the patients, some antibiotics offered at CLSI, and full funding were not available. Another limitation was the lack of testing for environmental samples, as S. maltophilia could be present in the hospital environment and on the equipment. By analyzing environmental samples and using molecular typing techniques, we were able to identify the source of the contamination. These limitations were overcome by the hospital administrators who engaged with laboratories and research centers.
Conclusion
The results of this study indicate that S. maltophilia may cause diseases such as bacteremia in hospitalized pediatric patients. Given the immunodeficiency in some hospitalized children, it is important to isolate S. maltophilia from samples taken from vulnerable pediatric population. Biofilm formation by S. maltophilia should be considered a major challenge in eliminating drug-resistant pathogens from hospitals. As of now, LEV and TMP/SMX can be considered effective antibiotics against S. maltophilia.
Disclosure
The authors declared no conflicts of interest. This research was funded by a grant from Hamadan University of Medical Sciences, Hamadan, Iran (Grant No: 9903131469).
Acknowledgments
The results described in this study are extracted from Hadis Divakan’s master’s thesis in medical microbiology, for which Leili Shokoohizadeh was a supervisor and Mohammad Yousef Alikhani an advisor. The authors would like to thank all members of microbiology laboratory of Hamadan University of Medical Science and the staff of Hamadan and Borujerd hospitals.
references
- 1. Curtis LT. Prevention of hospital-acquired infections: review of non-pharmacological interventions. J Hosp Infect 2008 Jul;69(3):204-219.
- 2. Lagamayo EN. Antimicrobial resistance in major pathogens of hospital-acquired pneumonia in Asian countries. Am J Infect Control 2008 May;36(4)(Suppl):S101-S108.
- 3. Juhász E, Pongrácz J, Iván M, Kristóf K. Antibiotic susceptibility of sulfamethoxazole-trimethoprim resistant Stenotrophomonas maltophilia strains isolated at a tertiary care centre in Hungary. Acta Microbiol Immunol Hung 2015 Sep;62(3):295-305.
- 4. Geng Y, Wang K, Chen D, Huang X, He M, Yin Z. Stenotrophomonas maltophilia, an emerging opportunist pathogen for cultured channel catfish, Ictalurus punctatus, in China. Aquacult Res 2010;308(3-4):132-135.
- 5. Naidu P, Smith S. A review of 11 years of Stenotrophomonas maltophilia blood isolates at a tertiary care institute in Canada. Can J Infect Dis Med Microbiol 2012;23(4):165-169.
- 6. Alsuhaibani M, Aljarbou A, Althawadi S, Alsweed A, Al-Hajjar S. Stenotrophomonas maltophilia bacteremia in children: risk factors and mortality rate. Antimicrob Resist Infect Control 2021 Jan;10(1):19.
- 7. Micozzi A, Venditti M, Monaco M, Friedrich A, Taglietti F, Santilli S, et al. Bacteremia due to Stenotrophomonas maltophilia in patients with hematologic malignancies. Clin Infect Dis 2000 Sep;31(3):705-711.
- 8. Sattler CA, Mason EO Jr, Kaplan SL. Nonrespiratory Stenotrophomonas maltophilia infection at a children’s hospital. Clin Infect Dis 2000 Dec;31(6):1321-1330.
- 9. Jeon YD, Jeong WY, Kim MH, Jung IY, Ahn MY, Ann HW, et al. Risk factors for mortality in patients with Stenotrophomonas maltophilia bacteremia. Medicine (Baltimore) 2016 Aug;95(31):e4375.
- 10. Wang YL, Scipione MR, Dubrovskaya Y, Papadopoulos J. Monotherapy with fluoroquinolone or trimethoprim-sulfamethoxazole for treatment of Stenotrophomonas maltophilia infections. Antimicrob Agents Chemother 2014;58(1):176-182.
- 11. Flores-Treviño S, Bocanegra-Ibarias P, Camacho-Ortiz A, Morfín-Otero R, Salazar-Sesatty HA, Garza-González E. Stenotrophomonas maltophilia biofilm: its role in infectious diseases. Expert Rev Anti Infect Ther 2019 Nov;17(11):877-893.
- 12. Gherardi G, Creti R, Pompilio A, Di Bonaventura G. An overview of various typing methods for clinical epidemiology of the emerging pathogen Stenotrophomonas maltophilia. Diagn Microbiol Infect Dis 2015 Mar;81(3):219-226.
- 13. Hu LF, Chen GS, Kong QX, Gao LP, Chen X, Ye Y, et al. Increase in the prevalence of resistance determinants to trimethoprim/sulfamethoxazole in clinical Stenotrophomonas maltophilia isolates in China. PLoS One 2016 Jun;11(6):e0157693.
- 14. CLSI. Performance standards for antimicrobial susceptibility testing. 31st ed. CLSI supplement M100. Clinical and Laboratory Standards Institute; 2021.
- 15. Di Bonaventura G, Spedicato I, D’Antonio D, Robuffo I, Piccolomini R. Biofilm formation by Stenotrophomonas maltophilia: modulation by quinolones, trimethoprim-sulfamethoxazole, and ceftazidime. Antimicrob Agents Chemother 2004 Jan;48(1):151-160.
- 16. Oliveira CF, Paim TG, Reiter KC, Rieger A, D’Azevedo PA. Evaluation of four different DNA extraction methods in coagulase-negative staphylococci clinical isolates. Rev Inst Med Trop Sao Paulo 2014 Jan-Feb;56(1):29-33.
- 17. Bostanghadiri N, Ghalavand Z, Fallah F, Yadegar A, Ardebili A, Tarashi S, et al. Characterization of phenotypic and genotypic diversity of Stenotrophomonas maltophilia strains isolated from selected hospitals in Iran. Front Microbiol 2019 May;10:1191.
- 18. Zarei O, Shokoohizadeh L, Hossainpour H, Alikhani MY. Molecular analysis of Pseudomonas aeruginosa isolated from clinical, environmental and cockroach sources by ERIC-PCR. BMC Res Notes 2018 Sep;11(1):668.
- 19. Hajiahmadi F, Safari N, Alijani P, Mordadi A, Arabestani MR. The frequency of integrons of antibiotic resistant in Stenotrophomonas maltophilia isolates in Hamadan/Iran. Iran J Med Microbiol 2016;10(4):10-16.
- 20. Baseri Z, Dehghan A, Yaghoubi S, Razavi S. Prevalence of resistance genes and antibiotic resistance profile among Stenotrophomonas maltophilia isolates from hospitalized patients in Iran. New Microbes New Infect 2021 Sep;44:100943.
- 21. Duan Z, Qin J, Li C, Ying C. Clinical and molecular epidemiology of Stenotrophomonas maltophilia in pediatric patients from a Chinese teaching hospital. Front Cell Infect Mic 2020:411.
- 22. Gayretli Aydιn ZG, Tanir G, Bayhan GI, Aydin Teke T, Metin Akçan O, Kaman A, et al. Risk factors of Stenotrophomonas maltophilia blood stream infections: comparison with other gram-negative blood stream infections in children. Pediatr Infect Dis J 2020 Dec;39(12):e406-e409.
- 23. Wu H, Wang JT, Shiau YR, Wang HY, Lauderdale TL, Chang SC; TSAR Hospitals. A multicenter surveillance of antimicrobial resistance on Stenotrophomonas maltophilia in Taiwan. J Microbiol Immunol Infect 2012 Apr;45(2):120-126.
- 24. Ebrahim-Saraie HS, Heidari H, Soltani B, Mardaneh J, Motamedifar M. Prevalence of antibiotic resistance and integrons, sul and Smqnr genes in clinical isolates of Stenotrophomonas maltophilia from a tertiary care hospital in Southwest Iran. Iran J Basic Med Sci 2019 Aug;22(8):872-877.
- 25. Malekan M, Tabaraie B, Akhoundtabar L, Afrough P, Behrouzi A. Distribution of class I integron and smqnr resistance gene among Stenotrophomonas maltophilia isolated from clinical samples in Iran. Avicenna J Med Biotechnol 2017 Jul-Sep;9(3):138-141.
- 26. Kaur P, Gautam V, Tewari R. Distribution of class 1 integrons, sul1 and sul2 genes among clinical isolates of Stenotrophomonas maltophilia from a tertiary care hospital in North India. Microb Drug Resist 2015 Aug;21(4):380-385.
- 27. Mansouri S, Razavi M, Norouzi F, Gholamhoseinian NS. Prevalence of β-Lactamase production and antimicrobial susceptibility of multidrug resistant clinical isolates of non-fermenting gram negative bacteria from hospitalized patients in Kerman/Iran. Jundishapur J Microbiol 2012;5(2):405-410.
- 28. Hu LF, Chen GS, Kong QX, Gao LP, Chen X, Ye Y, et al. Increase in the prevalence of resistance determinants to trimethoprim/sulfamethoxazole in clinical Stenotrophomonas maltophilia isolates in China. PLoS One 2016 Jun;11(6):e0157693.
- 29. Moriceau C, Eveillard M, Lemarié C, Chenouard R, Pailhoriès H, Kempf M. Stenotrophomonas maltophilia susceptibility to ceftazidime-avibactam combination versus ceftazidime alone. Med Mal Infect 2020 May;50(3):305-307.
- 30. Flores-Treviño S, Gutiérrez-Ferman JL, Morfín-Otero R, Rodríguez-Noriega E, Estrada-Rivadeneyra D, Rivas-Morales C, et al. Stenotrophomonas maltophilia in Mexico: antimicrobial resistance, biofilm formation and clonal diversity. J Med Microbiol 2014 Nov;63(Pt 11):1524-1530.
- 31. Zhuo C, Zhao QY, Xiao SN. The impact of spgM, rpfF, rmlA gene distribution on biofilm formation in Stenotrophomonas maltophilia. PLoS One 2014 Oct;9(10):e108409.
- 32. Corlouer C, Lamy B, Desroches M, Ramos-Vivas J, Mehiri-Zghal E, Lemenand O, et al; Collège de Bactériologie-Virologie-Hygiène des Hôpitaux de France. Stenotrophomonas maltophilia healthcare-associated infections: identification of two main pathogenic genetic backgrounds. J Hosp Infect 2017 Jun;96(2):183-188.
- 33. Valdezate S, Vindel A, Martín-Dávila P, Del Saz BS, Baquero F, Cantón R. High genetic diversity among Stenotrophomonas maltophilia strains despite their originating at a single hospital. J Clin Microbiol 2004 Feb;42(2):693-699.