Among the blood group systems, the Rhesus (Rh) system is the second in clinical importance after the ABO system. The Rh system has two main genes: RHD encodes the D antigen and RHCE encodes for C/c and E/e antigens both with 10 exons.1 RHD and RHCE genes each produce a protein antigen with 417 amino acids long. The most important antigens of the Rh system are D, C.c, E, and e.2 The immunogenicity of Rh antigens differs, with D antigen being the most immunogenic.3 To prevent alloimmunization due to anti-D, exposure of D-negative individuals to D-positive red blood cells should be avoided. Therefore, correct D phenotyping of donor’s red blood cells is essential to avoid such anti-D alloimmunization.
In most laboratories, serology is the method of choice to detect D antigen; however, it has limitations. Studies have shown that D variants such as weak D, Del phenotype, and partial D may be missed by standard serologic methods including the indirect antiglobulin test, and may cause anti-D immunization when transfused to D-negative recipients. Garratty estimated that the blood of at least 120 weak D or Del donors (though serologically type D negative) is transfused to D-negative recipients annually in Southern California.4 In another study on 46 133 serologically D-negative donors, the RHD genotyping showed that 96 samples had RHD gene, half of which harbored Del phenotype.5 Moussa et al,6 study realized that a partial D sample type DBT was mistyped as D-negative by serological tests. RHD gene molecular typing can overcome such limitations of serology.
The D-negative phenotype has a high molecular diversity which explains the discrepancies found between serologic and molecular methods.7 The frequency of D negative in Omanis is 8.35%,8 but the molecular background explaining this phenotype is still unknown in this population. With the aim to explore the molecular background of the possibility of serological D negative for any D variants, we describe an unusual case of serological D negative with the presence of an entire RHD gene in an Omani blood donor.
Case Report
A 43-year-old B RhD negative Omani male donor passed all eligibility tests and donated blood. Serological Rh phenotyping showed a D-C-c+E-e+ phenotype giving the initial impression of a possible dce/dce genotype. For molecular analysis, the presence of RHD exons 1 through 7 and RHD exons 9 and 10 was observed and found to be positive for all RHD exons except RHD exon 5. Sequencing of these RHD exons ruled out D variants. Sequencing of RHD intron 3/exon 4 for RHD pseudogen (RHDΨ) revealed 37 bp insertion with c.609 G>A mutation. This suggests and confirms the presence of the African RHD genotype responsible for the serological D-negative phenotype in this donor. The serological D negative was considered a true D negative with a possible Dce/dce genotype.
Discussion
The molecular background of D negative has been extensively studied in Caucasians with frequencies between 15% and 17% and Africans with frequencies between 1% and 7%.9–11 Two molecular backgrounds exist in D-negative Africans: RHDΨ and the RHD-CE-Ds hybrid gene that does not express D antigen but encodes an altered C antigen.12–14 In most Caucasians, the frequent cause for D negative phenotype is the lack of the entire RHD gene.15
In the present case, molecular analysis showed the presence of a complete RHD gene along with RHDΨ. This D negative is predicted to be either hemizygous or homozygous for RHDΨ gene. Omani populations are known to have an admixture of African genes,16 which can present a high variety of RHD alleles and explains the existence of RHDΨ.
RHDΨ is characterized by inactivation of D gene by insertion of 37 bp at the intron 3/exon 4 boundary of RHD gene that introduces a frameshift and translation termination. In addition, a nonsense (Tyr>stop) mutation in exon 6 causes premature termination of translated protein.12 RHDΨ associated nucleotides and amino acids changes related to wild-type RHD gene can be viewed in Figure 1. RHD gene deletion is a common cause of D negative in Africans, however around 67% are at least heterozygous to RHDΨ.17
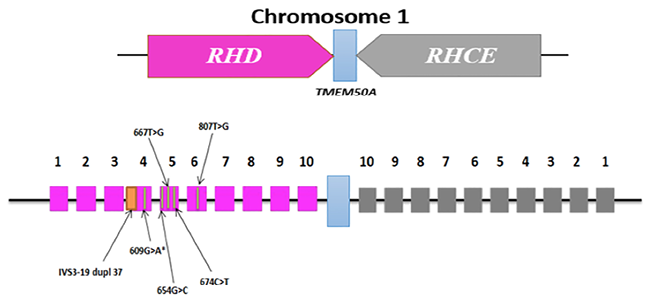 RHD and RHCE genes are separated by a small membrane protein TMEM50A gene. The numbers indicate the exon number on both RHD and RHCE genes. The pink boxes represent RHD exons and the grey boxes represent RHCE exons. The orange band within the pink box of RHD exon 4 represents a 37 bp insert which is a duplication of a sequence spanning the intron 3 (at -19 nucleotide sequence) – exon 4 boundary (IVS-19 dup 37). The green vertical lines represent point mutations associated with RHDΨ gene at RHD exon 4 (609G>A), RHD exon 5 (654G>C, 667T>G and 674C>T), and RHD exon 6 (807T>G). Asterisk (*) indicates a point mutation that does not result in an amino acid change. RHDΨ gene has no effect on RHCE gene.
RHD and RHCE genes are separated by a small membrane protein TMEM50A gene. The numbers indicate the exon number on both RHD and RHCE genes. The pink boxes represent RHD exons and the grey boxes represent RHCE exons. The orange band within the pink box of RHD exon 4 represents a 37 bp insert which is a duplication of a sequence spanning the intron 3 (at -19 nucleotide sequence) – exon 4 boundary (IVS-19 dup 37). The green vertical lines represent point mutations associated with RHDΨ gene at RHD exon 4 (609G>A), RHD exon 5 (654G>C, 667T>G and 674C>T), and RHD exon 6 (807T>G). Asterisk (*) indicates a point mutation that does not result in an amino acid change. RHDΨ gene has no effect on RHCE gene.
Figure 1: Schematic diagram on the molecular background of RHDΨ gene that gives D-negative phenotype.
In this case report, a previously described primer pair was used to amplify both wild-type RHD and RHD with a 37 bp insert specific for RHDΨ. The presence of 37 bp insertion was confirmed by Sanger sequencing. A previously described sequence-specific primer for RHD exon 5 was designed in a way so that the forward primer 3’ specific for wild type c.654 in exon 5 does not amplify mutation G>C (M218I) associated with RHDΨ.18 Therefore, amplification of RHD exon 4 and nonamplification of RHD exon 5 further confirmed the existence of RHDΨ. We were uncertain if the presence of RHDΨ is homozygous (RHDΨ/RHDΨ) or hemizygous (RHDΨ./del). D zygosity testing would have been helpful to unveil the same.
Conclusion
This is the first molecularly analyzed case that revealed African RHDΨ existence in an Omani blood donor. Our observation drives us to realize the necessity to study the molecular background of D-negative phenotype in Omanis. The molecular basis of D zygosity determination would be a good approach to further explore the case.
Disclosure
The authors declared no conflicts of interest. Informed written consent was taken from the patient.
references
- 1. Chérif-Zahar B, Mattéi MG, Le Van Kim C, Bailly P, Cartron JP, Colin Y. Localization of the human Rh blood group gene structure to chromosome region 1p34.3-1p36.1 by in situ hybridization. Hum Genet 1991 Feb;86(4):398-400.
- 2. Flegel WA. The genetics of the Rhesus blood group system. Blood Transfus 2007 Apr;5(2):50-57.
- 3. Harmening DM, Forneris G, Tubby BJ. Modern blood banking and transfusion practices. 6th ed. Philadelphia: FA Davis Company Publications; 2012. p. 149-165.
- 4. Engelfriet C, Reesink HW. Testing for weak D. Vox Sang 2006 Feb;90(2):140-153.
- 5. Flegel WA, von Zabern I, Wagner FF. Six years’ experience performing RHD genotyping to confirm D- red blood cell units in Germany for preventing anti-D immunizations. Transfusion 2009 Mar;49(3):465-471.
- 6. Moussa H, Tsochandaridis M, Chakroun T, Jridi S, Abdelneji B, Hmida S, et al. Molecular background of D-negative phenotype in the Tunisian population. Transfus Med 2012 Jun;22(3):192-198.
- 7. Sassi A, Ouchari M, Houissa B, Romdhane H, Abdelkefi S, Chakroun T, et al. RHD genotyping and its implication in transfusion practice. Transfus Apher Sci 2014 Dec;51(3):59-63.
- 8. Allawati M, Al-Kalbani L, Al-Balushi T. ABO and Rh (D) phenotypes, allele frequencies and estimated genotypes in Omanis: a retrospective study from armed forces hospital. JMSCR 2021;9(4):1-7.
- 9. Westhoff CM. The structure and function of the Rh antigen complex. Semin Hematol 2007 Jan;44(1):42-50.
- 10. Touinssi M, Chapel-Fernandes S, Granier T, Bokilo A, Bailly P, Chiaroni J. Molecular analysis of inactive and active RHD alleles in native Congolese cohorts. Transfusion 2009 Jul;49(7):1353-1360.
- 11. Weinstock C. It is worthwhile filling in the remaining blank spots for blood group antigen frequencies. Blood Transfus. 2014 Jan;12(1):3-6.
- 12. Singleton BK, Green CA, Avent ND, Martin PG, Smart E, Daka A, et al. The presence of an RHD pseudogene containing a 37 base pair duplication and a nonsense mutation in Africans with the Rh D-negative blood group phenotype. Blood 2000 Jan;95(1):12-18.
- 13. Faas BH, Beckers EA, Wildoer P, Ligthart PC, Overbeeke MA, Zondervan HA, et al. Molecular background of VS and weak C expression in blacks. Transfusion 1997 Jan;37(1):38-44.
- 14. Daniels GL, Faas BH, Green CA, Smart E, Maaskant-van Wijk PA, Avent ND, et al. The VS and V blood group polymorphisms in Africans: a serologic and molecular analysis. Transfusion 1998 Oct;38(10):951-958.
- 15. Wagner FF, Flegel WA. RHD gene deletion occurred in the Rhesus box. Blood 2000 Jun;95(12):3662-3668.
- 16. Al-Dughaishi T, Al Harrasi Y, Al-Duhli M, Al-Rubkhi I, Al-Riyami N, Al Riyami A, et al. Red cell alloimmunization to rhesus antigen among pregnant women attending a tertiary care hospital in Oman. Oman Med J 2016 Jan;31(1):77-80.
- 17. Daniels G. Variants of RhD–current testing and clinical consequences. Br J Haematol 2013 May;161(4):461-470.
- 18. Legler TJ, Liu Z, Mavrou A, Finning K, Hromadnikova I, Galbiati S, et al. Workshop report on the extraction of foetal DNA from maternal plasma. Prenat Diagn 2007 Sep;27(9):824-829.