Pertussis is a highly contagious respiratory infection caused by the bacterial species Bordetella pertussis.1 This disease accounts for significant morbidity and mortality among infants and children, particularly in low-income countries where immunization programs are not well established.2 In 2008, 195 000 children were estimated to die from the disease worldwide.3
Pertussis immunization has resulted in a significant drop in pertussis cases worldwide.2 However, the resurgence of cases was noticed during the last decades, even in countries with high vaccination coverage.4 In Oman, there has been a significant increase in the number of reported cases, rising from 56 cases in 2011 to 309 cases in 2013.5 Possible reasons may include incomplete effectiveness of the vaccine, waning vaccine-induced immunity, the adaptation of B. pertussis strains, increased awareness of the disease, and improved reporting.6 In addition, the atypical clinical presentation of pertussis seen in immunized individuals and adults makes the clinical diagnosis challenging, with the ongoing risk of transmission to vulnerable individuals.7 The resurgence in cases and the challenging clinical diagnosis of pertussis highlight the importance of using a rapid and accurate diagnostic method, leading to early treatment and the interruption of its transmission.
Culture is considered the gold standard for diagnosing pertussis because of its high specificity.8,9 However, studies have shown that it has suboptimal sensitivity ranging from 15% to 60%.10 It is also labor-intensive and may take up to six or seven days to finalize the results.
The most commonly used molecular method for the detection of B. pertussis is real-time polymerase chain reaction (PCR). Although previous studies have shown that PCR has superior sensitivity compared with culture, the specificity was variable among these studies. In addition, limited studies used clinical data to verify the specificity of PCR.11–13 This study aimed to evaluate real-time PCR for the diagnosis of pertussis, emphasizing the importance of clinical correlation in determining its specificity.
Methods
This retrospective diagnostic test accuracy study was conducted at Central Public Health Laboratories (CPHL), Ministry of Health, which receives samples from clinically suspected pertussis cases from all over Oman to perform diagnostic tests.
The study included nasopharyngeal/throat specimens sent in appropriate media (charcoal media) and tested by both culture and real-time PCR during the three-year study period: January 2014 to December 2016. The samples sent in viral transport media were tested only by PCR and thus were excluded from this study.
Culture was performed from nasopharyngeal/throat swabs transported in appropriate media supporting the viability of organisms (e.g., charcoal media) sent in a cool box. In the lab, the swabs were pre-warmed and then streaked in Bordetella selective agar (charcoal agar). A quality control sample was run for each batch of samples. The specimens were incubated at 37 °C in an ambient aerobic atmosphere for five days. The plates were inspected after 48 hours and onwards to check for growth. Any growth detected was tested for the presence of members of the genus Bordetella. The species B. pertussis was identified by its characteristic pearl-like colonies, Gram stain, oxidase test, and antisera.
In the real-time PCR method, the nasopharyngeal/throat samples were put in 2 mL of phosphate buffer saline which then underwent extraction according to the kit manufacturer’s instructions (Qiagen® DNeasy Blood & Tissue Kit, Germany). Amplification was then run using an in-house multiplex assay to target the IS 481, IS 1001, and hIS 1001 genes according to the Centers for Disease Control (CDC) protocol14 in addition to internal control. Targeting these genes enables real-time PCR to give a distinct positivity profile for each species of Bordetella such as B. pertussis, B. parapertussis, and B. holmesii. Positive IS 481 with negative IS 1001 and hIS 1001 indicates B. pertussis. Although uncommonly encountered, samples with positive IS 481 with high cycle threshold value (i.e., Ct ≥ 35) were further tested using a commercial CE marked FTD kit (Fast Track Diagnostics, Luxembourg) to confirm B. pertussis.
The data—lab number, date of sample collection, date of sample reception in CPHL, culture and real-time PCR results, and date of results release—were collected from laboratory records and entered in EpiData version 3.1. The analysis of the data was performed through IBM SPSS version 22 (IBM Corp. Released 2013. IBM SPSS Statistics for Windows, Version 22.0. Armonk, NY: IBM Corp.). Total turnaround time (TTAT; defined as the interval from sample collection to the reporting of results) and turnaround time (TAT; defined as the interval from sample receipt to the reporting of results) were calculated for both real-time PCR and culture. The transit time of the samples (from sample collection to sample reception in CPHL) was also calculated. In addition, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated for PCR in comparison to culture.
Cases that were pertussis-positive by real-time PCR but negative by culture were further assessed to rule out false-positive results. The clinical data were collected from the electronic health information system which connects all Ministry of Health hospitals. Demographic data, clinical data (including presence and duration of cough, presence of paroxysms of cough, inspiratory whoop, post-tussive vomiting, and apnea), presence or absence of alternative diagnoses, and epidemiologic links were collected in data collection sheets and entered in EpiData software before being exported to IBM SPSS for statistical analysis. The clinical data was compared against the clinical criteria of four different pertussis case definitions: CDC-2014, Canada-2009, Europe-2008, and Australia-2014.15
Ethical approval was obtained from Research and Ethical Review and Approve Committee, Ministry of Health, Sultanate of Oman, in August 2017, vide MoH/CSR/17/5902. All work in relation to the patients being reported anonymously, informed consent was not required.
Results
Out of the 907 nasopharyngeal/throat specimens received at CPHL during the three-year study period, 590 samples from 590 patients fulfilled the inclusion criteria (sent in charcoal media and kept at appropriate temperature) and were included in the study [Figure 1]. The demographic data of the 590 patients are shown in Table 1. The majority of the included samples (90.7%) were from children < 1 year of age.
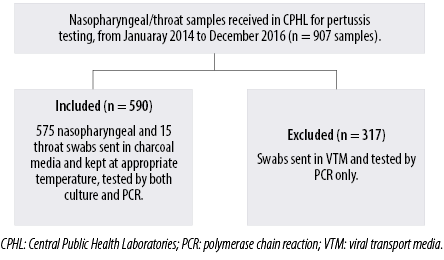
Figure 1: A flow chart showing the included samples in the study.
Table 1: Demographic data of the included patients.
|
Sex |
|
|
|
Male |
326 |
55.3 |
|
Female |
264 |
44.7 |
|
Total |
590 |
100 |
|
Nationality |
|
|
|
Omani |
586 |
99.3 |
|
Non-Omani |
4 |
0.7 |
|
Total |
590 |
100 |
|
Age, years |
|
|
|
< 1 |
535 |
90.7 |
|
1–5 |
36 |
6.1 |
|
6–15 |
9 |
1.5 |
|
16–50 |
8 |
1.4 |
|
> 50 |
2 |
0.3 |
|
Total |
590 |
100 |
|
Sample type |
|
|
|
Nasopharyngeal |
575 |
97.5 |
|
Throat |
15 |
2.5 |
Table 2: The number of positive and negative samples tested by real-time polymerase chain reaction (PCR) compared with those via culture.
|
Positive |
26 |
47 |
73 |
|
Negative |
0 |
517 |
517 |
Table 3: The clinical presentation of the 44 PCR–positive culture-negative cases of suspected pertussis.
|
Paroxysms of cough |
|
|
Present |
43 |
|
Absent |
0 |
|
Not mentioned |
1 |
|
Inspiratory whoop |
|
|
Present |
8 |
|
Absent |
0 |
|
Not mentioned |
36 |
|
Post-tussive vomiting |
|
|
Present |
21 |
|
Absent |
9 |
|
Not mentioned |
14 |
|
Apnea (if age is < 1 year) |
|
Present |
17 |
|
Absent |
13 |
|
Not mentioned |
14 |
|
Cough duration |
|
|
< 2 week |
22 |
|
≥ 2 weeks |
22 |
|
Epidemiologic link |
|
Present |
0 |
|
Absent |
2 |
Real-time PCR was positive for 73 out of the 590 samples (12.4%), while culture was positive for 26 (4.4%). All the 26 positive samples by culture were also positive by PCR, giving PCR a sensitivity of 100% [Table 2]. Out of the 564 negative samples by culture, 47 tested positive by PCR. The calculated specificity, PPV, and NPV for PCR compared with those for culture were 91.7%, 35.6%, and 100%, respectively.
The mean TAT was 3.4 days for real-time PCR compared with 6.2 days for culture. The transit time ranged between < 24 hours to 23 days, with an overall mean transit time of 2.3 days. The calculated mean of TTAT was 5.7 days for real-time PCR compared to 8.5 days for culture.
To overcome the issue of a less sensitive gold standard (culture) with the potential of overcalling false-positive results for the evaluated test (real-time PCR) – which may, in turn, affect the calculated specificity and PPV – further analysis was done for the PCR-positive culture-negative cases. A total of 44 out of the 47 PCR-positive culture-negative cases were further analyzed using four different published case definitions. Table 3 summarizes the clinical presentations of these 44 cases. The number of cases that met the clinical criteria according to the four case definitions are shown in Table 4. Adding PCR-positive results to the clinical data, 44 (100%) cases were classified as confirmed pertussis cases using the Canada-2009 and Australia-2014 case definitions. Applying the CDC-2014 and European-2008 case definitions, the numbers of confirmed cases were 21 (47.7%) cases and 41 (93.2%) cases, respectively. The calculation of specificity after the addition of these confirmed cases and the exclusion of three cases with unavailable clinical data yielded a higher specificity for PCR that ranges between 95.7% and 100% considering the different pertussis case definitions. The calculation considering the CDC-2014 definition is illustrated in Table 5.
An analysis per age for all the 73 PCR-positive samples, found that the majority of cases (67/73) were for children < 1 year of age, the majority of whom (55/73) were < 3 months of age.
Table 4: Classification of the 44 Positive-polymerase chain reaction (PCR) negative- culture cases according to different pertussis case definitions.
|
Cases that meet pertussis clinical criteria/evidence |
21 |
44* |
41 |
44 |
*Canada-2009 case definition has different clinical criteria for suspect and probable cases. 44 cases met the suspect case criteria, while 21 cases met the probable case criteria. With positive real-time PCR, all cases that met the suspect criteria are classified as confirmed according to this case definition.
Table 5: A comparison of polymerase chain reaction (PCR) cases with confirmed cases via either culture or the CDC 2014 case definition.
|
Positive |
47 ** |
23 *** |
70 |
|
Negative |
0 |
517 |
517 |
* Excluding the three cases (PCR-positive/culture-negative) with no access to clinical data. ** Forty-seven is the sum of the 26 cases positive via both culture and PCR as well as the 21 cases classified as confirmed using the CDC 2014 case definition. *** Twenty-three cases resulted from the subtraction of 21 confirmed cases per the CDC 2014 case definition and the three cases with no access to clinical data from the total 47 cases (initially considered possible false-positive PCR results).
Discussion
In this study, we evaluated the real-time PCR assay for the detection of B. pertussis in two steps: (1) the evaluation of performance compared with the culture which is the gold standard and (2) the evaluation of test specificity by clinical correlation of the discrepant results. The study included nasopharyngeal and throat samples. It is well-established that nasopharyngeal specimens give the best yield for B. pertussis, while throat swabs have unacceptably low rates of recovery.16 In our study, only 15 throat swabs were tested, and since all the samples underwent testing by both PCR and culture, they were included in our study as their results affected both arms of the study equally. Out of the 15 throat swabs, 14 were negative by both tests, while one was PCR-positive/culture-negative, and also met the pertussis clinical case definition.
Environmental contamination of clinical specimens in clinics and cross-contamination within laboratories are reported to be associated with false-positive PCR results and several pseudo-outbreaks of pertussis in recent years.8 The use of multiple targets in real-time PCR may improve specificity, as shown by Tatti et al,14 whose targets and protocol were followed in our laboratory.
Our results showed that real-time PCR has high sensitivity, reaching 100% and a specificity of 91.7% compared with culture which is congruent with previous studies.9–16 However, the low sensitivity of the gold standard (i.e. culture) could have affected the calculated specificity and PPV of real-time PCR since 47 samples were culture negative and PCR positive.
For this reason, the specificity of real-time PCR was further evaluated for those cases with discrepant PCR/culture results by carrying out clinical correlation using different case definitions. Most cases met the pertussis clinical criteria/evidence according to the Canada-2009 (100%), Australia-2014 (100%), and Europe-2008 (93.2%) case definitions. By adding positive PCR to the clinical criteria, these cases were considered confirmed. The CDC-2014 clinical case definition was an exception since only 21 of the 44 (47.7%) cases met its clinical criteria. A cough duration of at least two weeks was the only clinical criterion not met by most of the remaining cases (22/44). Although only 47.7% of the assessed cases met the CDC-2014 case definition, considering these cases as confirmed after adding positive PCR, the specificity of real-time PCR improved to 95.7%. Thus, considering the different mentioned case definitions, specificity improved to 95.7–100%. Considering that PCR is more sensitive in the first three weeks of illness,16 it is not unexpected that the cases that presented early and did not meet the clinical criterion of a minimum cough duration of two weeks were PCR-positive.
The accumulating evidence for the accuracy of PCR has led CDC to update its case definition, which was approved by the Council of State and Territorial Epidemiologists and went into effect on January 1, 2020.17 The current CDC-2020 case definition classifies PCR-positive cases with acute cough illness as confirmed, regardless of cough duration or presence of other pertussis symptoms. The updated CDC definition has resulted from increased confidence in the accuracy of PCR testing as a consequence of significant improvement in the quality of PCR testing, introduction of multiple target PCR, and minimization of contamination due to improved laboratory practice.18
Previous studies that investigated PCR and clinical data together have shown high specificity of PCR at 97–98%,12,13 which are in line with our results.
Our results emphasize the importance of clinical correlation in determining the PCR specificity, particularly in the absence of a sensitive gold standard, and demonstrate how a combination of laboratory and clinical data greatly increases the chance of early and accurate diagnosis, facilitating management at individual and epidemiological levels.
Depending on cultures for diagnosis requires viable bacterial load, to be present at testing stage. Factors that may lead to suboptimal culture sensitivity include the stage of the disease when the sample was taken, the vaccination status of the patient, the age of the patient, and the specimen quality.19 In addition, delays in transit and sample processing, and methods of culturing, all affect the viability of the organisms leading to suboptimal culture sensitivity. PCR is less likely to be affected by these factors since organism does not need to be viable to detect target genes. Culture is also labor-intensive and may take five to seven days to yield results, while PCR can be completed within one day. In our cases, however, the mean TAT for real-time PCR was longer (3.4 days) in our study due to capacity issues. However, it was clearly shorter than the culture TAT (6.2 days). Introducing PCR in regional hospitals in Oman will likely improve the TAT, and result in timely accurate diagnosis of pertussis.
Our results showed that real-time PCR is a sensitive, fairly specific, and rapid test for diagnosing B. pertussis. Although our results did not show additional benefits for culture in pertussis diagnosis compared with real-time PCR, culture might still be helpful for the surveillance of circulating strains and for performing antimicrobial susceptibility testing when clinically indicated.
Although Oman has a high vaccination coverage,5 a high number of pertussis cases are still reported, particularly among infants. In fact, adolescents and adults who have not received tetanus-diphtheria-pertussis (Tdap) booster vaccinations can become infected or re-infected as immunity from childhood vaccination and natural disease wanes. Atypical presentation with high potential of missing the diagnosis might be seen in this population.20 Such individuals may unwittingly transmit the infection to infants too young to be vaccinated and who are at the highest risk of severe complications and death. This makes an accurate and fast test to diagnose pertussis essential.
The strengths of our study include the large number of samples, the use of a three-target multiplex PCR rather than a singleplex PCR, and the clinical correlation done for the discrepant results. The study’s main limitation was its retrospective design of the study, which led to the unavailability of some clinical information required in the clinical correlation.
Conclusion
Real-time PCR is a highly sensitive and specific test for the diagnosis of B. pertussis. We recommend it to be part of the diagnostic tests for all suspected pertussis cases. Based on these results, we recommend setting up multiplex PCR diagnostic facilities in regional hospitals in Oman to enable timely and accurate diagnosis of pertussis.
Disclosure
The authors declare no conflicts of interest. No funding was received for this study. The abstract was presented as a poster in Oman Medical Specialty Board (OMSB) research and career day 2017/2018, and also published in OMJ (2018), Vol. 33, No. 1 under OMSB career and research forum 2018: Abstracts.
Acknowledgements
We thank the bacteriology section technologists at the Central Public Health Laboratories, Muscat, for their technical support.
references
- 1. Carbonetti NH. Bordetella pertussis: new concepts in pathogenesis and treatment. Curr Opin Infect Dis 2016 Jun;29(3):287-294.
- 2. Muloiwa R, Kagina BM, Engel ME, Hussey GD. The burden of pertussis in low- and middle-income countries since the inception of the Expanded Programme on Immunization (EPI) in 1974: a systematic review protocol. Syst Rev 2015 May;4(1):62.
- 3. Kilgore PE, Salim AM, Zervos MJ, Schmitt H-J. Pertussis: microbiology, disease, treatment, and prevention. Clin Microbiol Rev 2016 Jul;29(3):449-486.
- 4. Mooi FR, Van Der Maas NA, De Melker HE. Pertussis resurgence: waning immunity and pathogen adaptation - two sides of the same coin. Epidemiol Infect 2014 Apr;142(4):685-694.
- 5. Ministry of Health. Annual health reports 2015. Chapter 8: Health domains. Directorate General of Planning, Department of Information and Statistics 2015 [cited 2017 July 29]. Available from: https://moh.gov.om/en/web/statistics/-/20-31.
- 6. Lapidot R, Gill CJ. The pertussis resurgence: putting together the pieces of the puzzle. Trop Dis Travel Med Vaccines 2016 Dec;2(1):26.
- 7. Tozzi AE, Celentano LP, Ciofi degli Atti ML, Salmaso S. Diagnosis and management of pertussis. CMAJ 2005 Feb;172(4):509-515.
- 8. Lee AD, Cassiday PK, Pawloski LC, Tatti KM, Martin MD, Briere EC, et al; Clinical Validation Study Group. Clinical evaluation and validation of laboratory methods for the diagnosis of Bordetella pertussis infection: culture, polymerase chain reaction (PCR) and anti-pertussis toxin IgG serology (IgG-PT). PLoS One 2018 Apr;13(4):e0195979.
- 9. Dragsted DM, Dohn B, Madsen J, Jensen JS. Comparison of culture and PCR for detection of Bordetella pertussis and Bordetella parapertussis under routine laboratory conditions. J Med Microbiol 2004 Aug;53(Pt 8):749-754.
- 10. Ting TX, Hashim R, Ahmad N, Abdullah KH. Detection of Bordetella pertussis from clinical samples by culture and end-point PCR in Malaysian patients. Int J Bacteriol 2013;2013:324136.
- 11. Grimprel E, Bégué P, Anjak I, Betsou F, Guiso N. Comparison of polymerase chain reaction, culture, and western immunoblot serology for diagnosis of Bordetella pertussis infection. J Clin Microbiol 1993 Oct;31(10):2745-2750.
- 12. Loeffelholz MJ, Thompson CJ, Long KS, Gilchrist MJR. Comparison of PCR, culture, and direct fluorescent-antibody testing for detection of Bordetella pertussis. J Clini Microbiol 1999 Sep;37(9):2872-2876.
- 13. Chia JH, Su LH, Lin PY, Chiu CH, Kuo AJ, Sun CF, et al. Comparison of multiplex polymerase chain reaction, culture, and serology for the diagnosis of Bordetella pertussis infection. Chang Gung Med J 2004 Jun;27(6):408-415.
- 14. Tatti KM, Sparks KN, Boney KO, Tondella ML. Novel multitarget real-time PCR assay for rapid detection of Bordetella species in clinical specimens. J Clin Microbiol 2011 Dec;49(12):4059-4066.
- 15. Cherry JD, Tan T, Wirsing von König C-H, Forsyth KD, Thisyakorn U, Greenberg D, et al. Clinical definitions of pertussis: summary of a global pertussis initiative roundtable meeting, February 2011. Clin Infect Dis 2012 Jun;54(12):1756-1764.
- 16. Centers of Disease Control and Prevention (CDC). Best practices for healthcare professionals on the use of polymerase chain reaction (PCR) for diagnosing pertussis. [cited 2019 July 13]. Available from: https://www.cdc.gov/pertussis/clinical/diagnostic-testing/diagnosis-pcr-bestpractices.html.
- 17. Blain A. Skoff. T, Cassiday P, Tondilla ML, Acosta A. Chapter 10: Pertussis. Manual for the surveillance of vaccine-preventable diseases. Centres for Disease Control and Prevention. [cited 2021 October 15]. Available from: www.cdc.gov/vaccines/pubs/surv-manual/chpt10-pertussis.html.
- 18. CSTE. Revision to the case definition for national pertussis surveillance. CSTE position statement 19-ID-08: Atlanta, GA: CSTE; 2019 [cited 2021 October 15]. Available from: https://cdn.ymaws.com/www.cste.org/resource/resmgr/2019ps/final/19-ID-08_Pertussis_final_7.3.pdf.
- 19. van der Zee A, Schellekens JF, Mooi FR. Laboratory diagnosis of pertussis. Clin Microbiol Rev 2015 Oct;28(4):1005-1026.
- 20. Syed MA, Bana NF. Pertussis. A reemerging and an underreported infectious disease. Saudi Med J 2014 Oct;35(10):1181-1187.