Multiple sclerosis (MS) is a chronic and progressive disease of the central nervous system, affecting about two million individuals worldwide.1 It is an autoimmune-mediated demyelinating disorder of the central nervous system that has four clinical forms: secondary progressive (SPMS), relapsing-remitting (RRMS), primary progressive (PPMS), and progressive relapsing (PRMS).2 It usually affects people between the ages of 20 and 50 years old. It is diagnosed more often in women than men and is estimated to be the most common cause of neurological disability in young adults.1 Common early signs of MS include vision problems, pain spasm, tingling, numbness, weakness, fatigue, and cognitive impairments. With the progression of the disease, it can disrupt the daily activities of the affected individuals.3
For many years, MS was considered an autoimmune disorder, mainly of interest to immunologists. Researchers believed that the placement of viral antigens in the structure of the myelin sheath could cause the immune system to attack and eliminate the neuron.4 However, in the mid-1990s, it was recognized that a neurodegenerative process was responsible for MS development that was unresponsive to immunosuppression. The neuropathology of MS emphasized the axonal changes followed by demyelination, which leads to neural cell death and disease progression.5 Autoreactive pathogenic helper T cells play a prominent role in the pathogenesis of MS.6
The pathogenesis of MS is best described by a multifactorial model incorporating interactions between genetic and environmental factors, including nutrition, climate, and infection.7,8 The role of genetic factors is increasingly taken into account, as the environmental effects are also largely influenced by the genetic characteristics of individuals.
The primary efforts to identify susceptibility genes were performed through linkage analysis in families with a high prevalence of the disease. Although the studies failed to find a prominent genetic association, the human leukocyte antigen region was identified to have a very strong association with MS.9 The first genome-wide association study (GWAS) in MS was carried out by the International Multiple Sclerosis Genetics Consortium (IMSGC), and several GWASs and candidate gene association studies provided evidence for the association of more than 200 susceptibility variants with MS.1
For example, a 2014 study demonstrated a significant association between the ApaI polymorphism in the vitamin D receptor gene and MS risk in the homozygous and codominant models.10 Another study in the same year showed a link between polymorphism in the interleukin (IL-2) receptor alpha gene and MS.11 In 2015, another study investigated the association between 3061AG polymorphism located in the very-late antigen 4 gene and the risk of MS,12 and more recently the authors of one study suggested that the aryl hydrocarbon receptor nuclear translocator-like rs3789327 CC genotype is associated with a higher risk for MS.13
The main goal of our study was to investigate the association of rs12487066 (CBLB), rs12044852 (CD58), rs10735781 (EVI5), rs3135388 (HLA), rs6897932 (IL7R), rs1321172 (PDE4B), rs10492972 (KIF1B), and rs9657904 (CBLB) polymorphisms with MS in the Iranian population. The results of this study can, in addition to, confirm or reject the results of previous studies, consider the importance of the examined polymorphisms as biomarkers for better diagnosis of MS.
Methods
The case group consisted of 83 patients (82.0% females and 18.0% males) with MS referred to Loghman Hospital in Tehran, Iran. Moreover, in this study, all procedures were performed on human participants.
Two experienced neurologists confirmed MS in all cases. All patients were also evaluated with McDonald criteria. The McDonald criteria maintained a scheme for diagnosing MS-based solely on clinical grounds and try to prove the existence of demyelinating lesions.14 According to their reports, as well as clinical evaluations by relevant specialists, patients did not experience any other neurological or mental disorder. Besides the case group, 100 physically and mentally healthy subjects (81.0% female and 19.0% male) were selected as a control group. None of the cases and controls had a drug addiction or alcohol abuse. The mean age of the case group was 35.2±8.6 years, and the mean age of the control group was 40.4±6.4 years. All subjects participated in the study with informed consent. The ethical committee of the Taban Genetics Center, Tehran, Iran, approved this study.
A 5 mL blood sample was taken from each case and control patient and transferred to the lab for genetic analysis in an EDTA tube. DNA isolation was performed using GENET BIOkit (Global Gene Network, Korea) based on the company protocol. The quantity and quality of DNA were evaluated by the Denovix NanoDrop device (Model Ds-11, USA) and gel electrophoresis, respectively. Amplification-refractory mutation system in conjunction with polymerase chain reaction (tetra-primer ARMS-PCR) was used to genotype the desired polymorphisms, rs12487066, rs12044852, rs10735781, rs3135388, rs6897932, rs1321172, rs10492972, and rs9657904. Using four specifically designed primers, tetra-primer ARMS-PCR allows the amplification of sample DNA only when it contains the target variant.
The tetra-primer ARMS-PCR was carried out in a final volume of 25 µL comprising 1 µL of the DNA sample, 2.5 µL of each outer primer, 2.5 µL of the forward inner primer, 2.5 µL of the reverse inner primer, 0.5 µL dNTPs, 1 µL MgCl2, 2.5 µL Buffer and 0.2 µL of TaDNA polymerase (Sinaclon, Iran). Amplicons were analyzed by 1% agarose gel electrophoresis in TAE 0.5 Xbuffer (Sinaclon, Iran), containing SYBR safe stain. Also, 10% of the samples were sequenced to ensure the amplification of the target regions. All of the primer sets used in this study were designed using the Primer 1 online tool (primer1.soton.ac.uk/primer1.html) and are shown in Table 1.
Table 1: All of the primers used in this study.
|
rs12487066 |
CBLB |
FO: TTTTCTACTATTGGGTACCCAGAGC
RO: CTTTTGTGGACTTCTTCCTCCC
FI: CAAGAAAAACTTAACGACTAAAAGTGCTT
RI: TTTGCTACAGCACCTTGTCAGTTAG |
|
rs12044852 |
CD58 |
FO: CAAGGAAGTCATGCTGGAACTGACAT
RO: CATGGACTTCATTGCTACAGCATTGA
FI: GGATACACACGTGATTCCTAACGGC
RI: CCCTTGCCCTCCTCATTCCAT |
|
rs10735781 |
EVI5 |
FO: AAATGCTAACAGAATTCATCAC
RO: CTTGTCTTTTTTCACTGTTGTT
FI: CAGACAAAAGTATAAAACTTACTGC
RI: GGATCATCCTTTTTGTAACAC |
|
rs3135388 |
HLA |
FO: GTCTAACAGAATGGGTAAGGCCAGTCTT
RO: GGTCCTGGGGAATATATGTGATCCTTT
FI: CAGTAGAGATCTCCCAACAAACCCAC
RI: GTCCTCATCAGGAAAACCTAAAGTGTGA |
|
rs6897932 |
IL7RA |
FO: CCCTCCATAAAGCTGTCAAATATGTC
RO: CAATAAATGGGGCTTAAGCTCTGACT
FI: AGGGGAGATGGATCCTATCTTACTCAT
RI: GAGAAAAAACTCAAAATGCTGAGGG |
|
rs1321172 |
PDE4B |
FO: ATCCTATTGAGCGGGGCTCTCAAATTT
RO: GTGCAAGAGAATGCAAAAAGAAGTGAA
FI: GATTCTCTGCTCCACTAAGGAGTAACTGC
RI: CTCACTCTCTTGCTCATTGCAAATCTC |
|
rs9657904 |
CBLB |
FO: ACTAACTTGTACCACTGCATCTTCCTC
RO: CCAAAATGTATGATAGGACCTTCAGTTG
FI: TTTCAAGTAGCTAAGGCTGAACTAATCCT
RI: TTGTTCTTTTTTTTTTTTTTATGAGTGGG |
SNPs: single nucleotide polymorphisms; FO: forward outer primer; RO: reverse outer primer; FI: forward inner primer; RI: reverse inner primer.
Table 2: PCR temperature protocol.
|
HLA (rs3135388) |
61o- 30 s |
|
IL7RA (rs6897932) |
59o- 30 s |
|
KIF1B (rs10492972) |
60o- 30 s |
|
EVI5 (rs10735781) |
59o- 30 s |
|
CD58 (rs12044852) |
62o- 30 s |
|
CBLB (rs12487066) |
58o- 30 s |
|
PDE4B (rs1321172) |
61o- 30 s |
PCR: polymerase chain reaction.
Table 3: The allele frequency of all examined polymorphisms.
|
rs10735781 |
C |
73 (88.0) |
95 (95.0) |
2.99, 0.080 |
|
G |
10 (12.0) |
5 (5.0) |
|
rs6897932 |
C |
43 (51.8) |
65 (65.0) |
3.26, 0.070 |
|
T |
40 (48.2) |
35 (35.0) |
|
rs12044852 |
C |
72 (86.7) |
88 (88.0) |
0.06, 0.790 |
|
A |
11 (13.3) |
12 (12.0) |
|
rs1321172 |
G |
56 (67.5) |
68 (68.0) |
0.005, 0.930 |
|
C |
27 (32.5) |
32 (32.0) |
|
rs12487066 |
C |
29 (34.9) |
45 (45.0) |
1.90, 0.160 |
|
T |
54 (65.1) |
55 (55.0) |
|
rs3135388 |
C |
60 (72.3) |
78 (78.0) |
0.79, 0.370 |
|
T |
23 (27.7) |
22 (22.0) |
|
T |
71 (85.5) |
86 (86.0) |
SNPs: single nucleotide polymorphisms.
Table 4: Genetic analysis results based on the multiple sclerosis model.
|
rs10735781 |
|
|
Codominant |
C/C |
64 (77.1) |
91 (91.0) |
1.00 |
0.029 |
|
C/G |
18 (21.7) |
8 (8.0) |
0.31 (0.13–0.76) |
|
G/G |
1 (1.2) |
1 (1.0) |
0.70 (0.04–11.45) |
|
Dominant |
C/C |
64 (77.1) |
91 (91.0) |
1.00 |
0.009 |
|
C/G-G/G |
19 (22.9) |
9 (9.0) |
0.33 (0.14–0.78) |
|
Recessive |
C/C-C/G |
82 (98.8) |
99 (99.0) |
1.00 |
0.890 |
|
G/G |
1 (1.2) |
1 (1.0) |
0.83 (0.05–13.45) |
|
Overdominant |
C/C-G/G |
65 (78.3) |
92 (92.0) |
1.00 |
0.008 |
|
C/G |
18 (21.7) |
8 (8.0) |
0.31 (0.13–0.77) |
|
Log-additive |
- |
- |
- |
0.40 (0.18–0.87) |
0.016 |
|
rs6897932 |
|
Codominant |
C/C |
22 (26.5) |
43 (43.0) |
1.00 |
0.012 |
|
C/T |
40 (48.2) |
46 (46.0) |
0.59 (0.30–1.15) |
|
T/T |
21 (25.3) |
11 (11.0) |
0.27 (0.11–0.65) |
|
Dominant |
C/C |
22 (26.5) |
43 (43.0) |
1.00 |
0.019 |
|
C/T-T/T |
61 (73.5) |
57 (57.0) |
0.48 (0.26–0.90) |
|
Recessive |
C/C-C/T |
62 (74.7) |
89 (89.0) |
1.00 |
0.011 |
|
T/T |
21 (25.3) |
11 (11.0) |
0.36 (0.16–0.81) |
|
Overdominant |
C/C-T/T |
43 (51.8) |
54 (54.0) |
1.00 |
0.770 |
|
C/T |
40 (48.2) |
46 (46.0) |
0.92 (0.51–1.64) |
|
Log-additive |
- |
- |
- |
0.53 (0.34–0.81) |
0.003 |
|
rs12044852 |
|
Codominant |
C/C |
63 (75.9) |
75 (75.0) |
1.00 |
0.980 |
|
C/A |
19 (22.9) |
24 (24.0) |
1.06 (0.53–2.11) |
|
A/A |
1 (1.2) |
1 (1.0) |
0.84 (0.05–13.70) |
|
Dominant |
C/C |
63 (75.9) |
75 (75.0) |
1.00 |
0.890 |
|
C/A-A/A |
20 (24.1) |
25 (25.0) |
1.05 (0.53–2.07) |
|
Recessive |
C/C-C/A |
82 (98.8) |
99 (99.0) |
1.00 |
0.890 |
|
A/A |
1 (1.2) |
1 (1.0) |
0.83 (0.05–13.45) |
|
Overdominant |
C/C-A/A |
64 (77.1) |
76 (76.0) |
1.00 |
0.860 |
|
C/A |
19 (22.9) |
24 (24.0) |
1.06 (0.53–2.12) |
|
Log-additive |
- |
- |
- |
1.03 (0.55–1.94) |
0.920 |
|
rs1321172 |
|
Codominant |
G/G |
39 (47.0) |
47 (47.0) |
1.00 |
1.000 |
|
C/G |
36 (43.4) |
43 (43.0) |
0.99 (0.54–1.83) |
|
C/C |
8 (9.6) |
10 (10.0) |
1.04 (0.37–2.88) |
|
Dominant |
G/G |
39 (47.0) |
47 (47.0) |
1.00 |
1.000 |
|
C/G-C/C |
44 (53.0) |
53 (53.0) |
1.00 (0.56–1.79) |
|
Recessive |
G/G-C/G |
75 (90.4) |
90 (90.0) |
1.00 |
0.930 |
|
C/C |
8 (9.6) |
10 (10.0) |
1.04 (0.39–2.77) |
|
Overdominant |
G/G-C/C |
47 (56.6) |
57 (57.0) |
1.00 |
0.960 |
|
C/G |
36 (43.4) |
43 (43.0) |
0.98 (0.55–1.77) |
|
Log-additive |
- |
- |
- |
1.01 (0.65–1.57) |
0.970 |
|
rs12487066 |
|
Codominant |
T/T |
35 (42.2) |
30 (30.0) |
1.00 |
0.150 |
|
C/T |
38 (45.8) |
50 (50.0) |
1.54 (0.81–2.93) |
|
C/C |
10 (12.1) |
20 (20.0) |
2.33 (0.95–5.75) |
|
Dominant |
T/T |
35 (42.2) |
30 (30.0) |
1.00 |
0.087 |
|
C/T-C/C |
48 (57.8) |
70 (70.0) |
1.70 (0.92–3.13) |
|
Recessive |
T/T-C/T |
73 (88) |
80 (80.0) |
1.00 |
0.140 |
|
C/C |
10 (12.1) |
20 (20.0) |
1.82 (0.80–4.15) |
|
Overdominant |
T/T-C/C |
45 (54.2) |
50 (50.0) |
1.00 |
0.570 |
|
C/T |
38 (45.8) |
50 (50.0) |
1.18 (0.66–2.12) |
|
Log-additive |
- |
- |
- |
1.53 (0.99–2.35) |
0.050 |
|
rs3135388 |
|
Codominant |
C/C |
43 (51.8) |
60 (60.0) |
1.00 |
0.330 |
|
C/T |
33 (39.8) |
36 (36.0) |
0.78 (0.42–1.44) |
|
T/T |
7 (8.4) |
4 (4.0) |
0.41 (0.11–1.49) |
|
Dominant |
C/C |
43 (51.8) |
60 (60.0) |
1.00 |
0.270 |
|
C/T-T/T |
40 (48.2) |
40 (40.0) |
0.72 (0.40–1.29) |
|
Recessive |
C/C-C/T |
76 (91.6) |
96 (96.0) |
1.00 |
0.210 |
|
T/T |
7 (8.4) |
4 (4.0) |
0.45 (0.13–1.60) |
|
Overdominant |
C/C-T/T |
50 (60.2) |
64 (64.0) |
1.00 |
0.600 |
|
C/T |
33 (39.8) |
36 (36.0) |
0.85 (0.47–1.55) |
|
Log-additive |
- |
- |
- |
0.71 (0.44–1.15) |
0.160 |
|
rs9657904 |
|
Codominant |
T/T |
60 (72.3) |
72 (72.0) |
1.00 |
0.970 |
|
C/T |
21 (25.3) |
25 (25.0) |
0.99 (0.51–1.95) |
|
C/C |
2 (2.4) |
3 (3.0) |
1.25 (0.20–7.73) |
|
Dominant |
T/T |
60 (72.3) |
72 (72.0) |
1.00 |
0.970 |
|
C/T-C/C |
23 (27.7) |
28 (28.0) |
1.01 (0.53–1.94) |
|
Recessive |
T/T-C/T |
81 (97.6) |
97 (97.0) |
1.00 |
0.810 |
|
C/C |
2 (2.4) |
3 (3.0) |
1.25 (0.20–7.68) |
|
Overdominant |
T/T-C/C |
62 (74.7) |
75 (75.0) |
1.00 |
0.960 |
|
C/T |
21 (25.3) |
25 (25.0) |
0.98 (0.50–1.92) |
OR: odds ratio; CI: confidence interval.
The initial denaturation cycle of PCR was carried out at 95 oC for 5 minutes for all samples. The amplification of all samples was performed through 30 cycles including denaturation at 95 oC for 30 seconds, annealing [Table 2], and extension at 72 oC for 30 seconds, following by a final extension step at 72 oC for 10 minutes.
The validity of the design of the primer sets was checked by NCBI BLAST before synthesis (Sinaclon, Iran).
We used central and dispersion indices to describe the results statistically. We used the Shapiro-Wilks test to test for normality. To compare the mean age between the two groups, the independent t-test or ’Wilcoxon’s nonparametric test was used, based on how data were distributed. The associations between polymorphisms and the disease were studied based on the codominant, dominant, recessive, and overdominant models. All statistical analyses were performed using the IBM SPSS Statistics 25 software (IBM Corp. Released 2017. IBM SPSS Statistics for Windows, Version 25.0. Armonk, NY: IBM Corp.).

Figure 1: The mean frequency of symptoms associated with multiple sclerosis in patients.
Results
We found no significant difference between the case and the control group in terms of the male/female ratio (χ = 0.03, p = 0.870). The assessment of the symptoms of the disease indicated that the most common symptoms included sleep disorders, difficulty in maintaining balance, and feeling exhausted [Figure 1].
Independent t-test showed that the mean age of the two groups were significantly different (t = -4.54, p < 0.001). Statistical analysis showed that the allele frequencies were not significantly different between the two groups [Table 3].
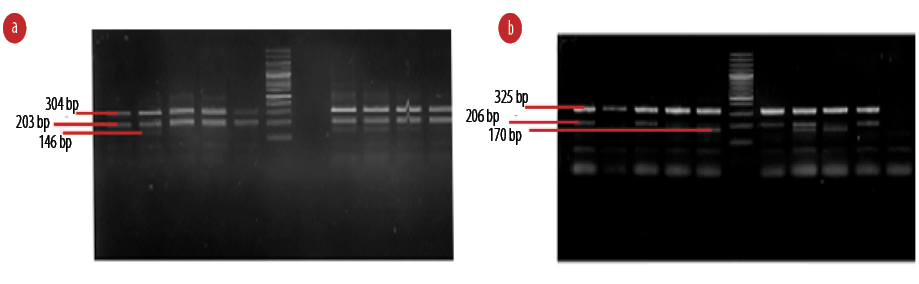
Figure 2: The amplified sequences of (a) rs10735781 and (b) rs6897932 through the tetra-primer amplification-refractory mutation system in conjunction with polymerase chain reaction method.
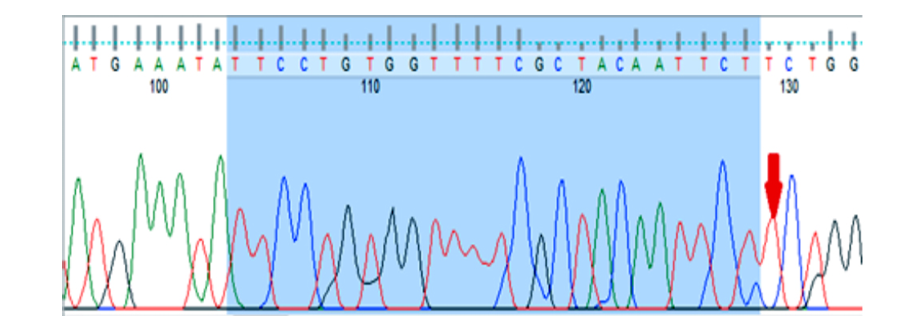
Figure 3: Image of gene sequencing KIF1B gene (rs10492972). This image related to homozygous normal genotype (TT).
Genetic analysis demonstrated that rs10735781 polymorphism was associated with MS codominantly (p = 0.029), overdominantly (p = 0.008), and dominantly (p = 0.009), and considering the significance levels the overdominant association seemed to be the preferred model [Table 4]. The rs6897932 polymorphism was found to be codominantly (p = 0.012), dominantly (p = 0.019), and recessively (p = 0.011) associated with the disease, and considering the significance levels, the recessive association was the preferred model. The rest of the polymorphisms did not show any significant association with the disease [Table 4]. The amplified sequences of the two polymorphisms are shown in Figure 2. The rs10492972 polymorphism genotype was similar in all subjects (TT) and, therefore, it was not analyzed. The amplified sequence of rs10492972 is shown in Figure 3.
Discussion
We investigated the association between rs12487066, rs12044852, rs10735781, rs3135388, rs6897932, rs1321172, rs10492972, and rs9657904 polymorphisms and MS in an Iranian population. Our results demonstrated that the rs10735781 polymorphism is overdominantly associated with the disease. Additionally, rs6897932 polymorphism is codominantly (p < 0.050) and recessively (p < 0.050) associated with the disease. No significant association was found between other polymorphisms and the disease.
The minor allele frequency of rs10735781 polymorphism in the general population is reported to be about 0.35,15 while in our study, it was 0.12 in the case group and 0.05 in the control group. Although achieving a precise conclusion requires studying larger populations, it could be inferred that the minor allele is associated with certain health conditions. This polymorphism is located at the EVI5 gene, which regulates cell cycle progression and cytokines by stabilizing the F-Box gene product.15
A 2008 study demonstrated a significant association between EVI5 and MS.16 The authors of the study further validated the risk effect of EVI5 for the general MS population in an independent set of 1318 MS patients. Although a more recent study found no significant association of EVI5 with MS,15 our study showed a significant association between this gene and the disease. Perhaps the main reason for the mentioned contradictions is the difference in the size and ethnicity of the study samples.
It is still ambiguous how the rs10735781 polymorphism plays a role in the pathology of the disease. It remains to be investigated how and to what extent it could influence T cell lymphomagenesis and function. Functional immunological studies stratified according to EVI5 genotype will clarify the role of this gene in MS pathology.
The minor allele frequency of rs6897932 in the general population is reported to be 0.17,17 while in our study it was 0.40 in the case group and 0.35 in the control group. Both of which have a great deal of difference with the general population.17
One of the most important hypotheses about the incidence of MS is the hypothesis of autoimmunity, which suggests that MS may be an autoimmune disease. Furthermore, most current MS therapies are based on drugs that attenuate the immune response.18 Therefore, it is not unlikely that genetic abnormalities in immune system cells are involved in MS pathology.
Some studies have confirmed that variation within the major histocompatibility complex (MHC) exerts a great risk in MS incidence.19 Furthermore, using the ImmunoChip genotyping assay, a study by the IMSGC found 103 discrete loci outside of the MHC region.20
There is a significant association between genetic variation in the IL7R gene and patients with progressive MS.21 More recently, an epistatic interaction that controls the IL7R splicing and increases MS risk was identified.22 Additionally, three IL7RA loci (rs3194051, rs987107, and rs11567686) are thought to be significantly associated with increasing MS risk.23 Our findings are in line with previous studies.23
Although the association of rs6897932 polymorphism with MS has not been investigated, its association with other autoimmune disorders, such as systemic lupus erythematosus,24 and asthma has been established.25 It is believed that rs6897932 polymorphism regulates monocyte surface IL7R induction by lipopolysaccharides and tumor necrosis factor stimulation.26
On the other hand, the associations of rs12487066,27 rs3135388,28 rs6897932,29 rs1321172,30 rs9657904, and rs1049297231 polymorphisms with MS have been found in previous studies, but we did not find such associations. The reason for this difference can be related to other risk factors. For example, that ethnicity,32 geographical coordinates,33 and family history34 can affect the risk of MS. Also, differences in the sample size and the male/female ratio can also lead to differences in results.
Conclusion
MS is a multifactorial disease, thus determining the genetic polymorphisms of genes might be an essential strategy as a marker of genetic predisposition to MS. This study is illustrated the association between the two polymorphisms, rs10735781 and rs6897932 on EVI5 and IL7RA genes, respectively, with increased MS in the Iranian population. EVI5 is mainly involved in cell cycle regulation and cytokinesis, while IL7RA predominantly acts as an immune response mediator, and their role in MS has been somewhat identified in previous studies. Thus, these results suggest that evaluation of EVI5 and IL7RA genetic polymorphisms could be considered a prognostic mechanism to identify individuals at higher risk of developing MS. However, structural and functional genetic studies in the future on larger populations can better capture the importance of these two genes in the onset of the disease.
Disclosure
The authors declared no conflicts of interest. No funding was received for this study.
Acknowledgements
This research was supported by the National Institute of Genetic Engineering and Biotechnology (NIGEB). We thank our colleagues who provided insight and expertise that greatly assisted the research.
references
- 1. Baranzini SE, Oksenberg JR. The genetics of multiple sclerosis: from 0 to 200 in 50 years. Trends Genet 2017 Dec;33(12):960-970.
- 2. Jafarzadeh A, Mahdavi R, Jamali M, Hajghani H, Nemati M, Ebrahimi H-A. Increased concentrations of interleukin-33 in the serum and cerebrospinal fluid of patients with multiple sclerosis. Oman Med J 2016 Jan;31(1):40-45.
- 3. Kister I, Bacon TE, Chamot E, Salter AR, Cutter GR, Kalina JT, et al. Natural history of multiple sclerosis symptoms. Int J MS Care 2013;15(3):146-158.
- 4. Chaudhuri A, Behan PO. Multiple sclerosis is not an autoimmune disease. Arch Neurol 2004 Oct;61(10):1610-1612.
- 5. Hauser SL, Oksenberg JR. The neurobiology of multiple sclerosis: genes, inflammation, and neurodegeneration. Neuron 2006 Oct;52(1):61-76.
- 6. Haghmorad D, Mahmoudi MB, Mahmoudi M, Rab SZ, Rastin M, Shegarfi H, et al. Calcium intervention ameliorates experimental model of multiple sclerosis. Oman Med J 2014 May;29(3):185-189.
- 7. Hedström AK, Alfredsson L, Olsson T. Environmental factors and their interactions with risk genotypes in MS susceptibility. Curr Opin Neurol 2016 Jun;29(3):293-298.
- 8. Olsson T, Barcellos LF, Alfredsson L. Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat Rev Neurol 2017 Jan;13(1):25-36.
- 9. Ligers A, Dyment DA, Willer CJ, Sadovnick AD, Ebers G, Risch N, et al; Canadian Collaborative Study Groups. Evidence of linkage with HLA-DR in DRB1*15-negative families with multiple sclerosis. Am J Hum Genet 2001 Oct;69(4):900-903.
- 10. Tizaoui K, Kaabachi W, Hamzaoui A, Hamzaoui K. Association between vitamin D receptor polymorphisms and multiple sclerosis: systematic review and meta-analysis of case-control studies. Cell Mol Immunol 2015 Mar;12(2):243-252.
- 11. Hartmann FJ, Khademi M, Aram J, Ammann S, Kockum I, Constantinescu C, et al. Multiple sclerosis-associated IL2RA polymorphism controls GM-CSF production in human TH cells. Nat Commun 2014 Oct;5(1):5056.
- 12. Ďurmanová V, Shawkatová I, Javor J, Párnická Z, Čopíková-Cundráková D, Turčáni P, et al. VLA4 gene polymorphism and susceptibility to multiple sclerosis in Slovaks. Folia Biol (Praha) 2015;61(1):8-13.
- 13. Lavtar P, Rudolf G, Maver A, Hodžić A, Starčević Čizmarević N, Živković M, et al. Association of circadian rhythm genes ARNTL/BMAL1 and CLOCK with multiple sclerosis. PLoS One 2018 Jan;13(1):e0190601.
- 14. Ntranos A, Lublin F. Diagnostic criteria, classification and treatment goals in multiple sclerosis: the chronicles of time and space. Curr Neurol Neurosci Rep 2016 Oct;16(10):90.
- 15. Mazdeh M, Rahimi M, Eftekharian MM, Omrani MD, Sayad A, Taheri M, et al. Ecotropic viral integration site 5 (EVI5) expression analysis in multiple sclerosis patients. Hum Antibodies 2017;26(3):113-119.
- 16. Hoppenbrouwers IA, Aulchenko YS, Ebers GC, Ramagopalan SV, Oostra BA, van Duijn CM, et al. EVI5 is a risk gene for multiple sclerosis. Genes Immun 2008 Jun;9(4):334-337.
- 17. Deelen J, Beekman M, Capri M, Franceschi C, Slagboom PE. Identifying the genomic determinants of aging and longevity in human population studies: progress and challenges. Bioessays 2013 Apr;35(4):386-396.
- 18. Gulcher JR, Vartanian T, Stefansson K. Is multiple sclerosis an autoimmune disease? Clin Neurosci 1994;2(3-4):246-252.
- 19. Sawcer S, Hellenthal G, Pirinen M, Spencer CC, Patsopoulos NA, Moutsianas L, et al; International Multiple Sclerosis Genetics Consortium; Wellcome Trust Case Control Consortium 2. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011 Aug;476(7359):214-219.
- 20. Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C, et al; International Multiple Sclerosis Genetics Consortium (IMSGC); Wellcome Trust Case Control Consortium 2 (WTCCC2); International IBD Genetics Consortium (IIBDGC). Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet 2013 Nov;45(11):1353-1360.
- 21. Traboulsee AL, Bernales CQ, Ross JP, Lee JD, Sadovnick AD, Vilariño-Güell C. Genetic variants in IL2RA and IL7R affect multiple sclerosis disease risk and progression. Neurogenetics 2014 Aug;15(3):165-169.
- 22. Galarza-Muñoz G, Briggs FB, Evsyukova I, Schott-Lerner G, Kennedy EM, Nyanhete T, et al. Human epistatic interaction controls IL7R splicing and increases multiple sclerosis risk. Cell 2017 Mar;169(1):72-84.e13.
- 23. Liu H, Huang J, Dou M, Liu Y, Xiao B, Liu X, et al. Variants in the IL7RA gene confer susceptibility to multiple sclerosis in Caucasians: evidence based on 9734 cases and 10436 controls. Sci Rep 2017 Apr;7(1):1207.
- 24. Wang XS, Wen PF, Zhang M, Hu LF, Ni J, Qiu LJ, et al. Interleukin-7 receptor single nucleotide polymorphism rs6897932 (C/T) and the susceptibility to systemic lupus erythematosus. Inflammation 2014 Apr;37(2):615-620.
- 25. Sinha S, Singh J, Jindal SK. Association of interleukin 7 receptor (rs1494555 and rs6897932) gene polymorphisms with asthma in a north Indian population. Allergy Rhinol (Providence) 2015 Jan;6(3):168-176.
- 26. Al-Mossawi H, Lau E, Danielli S, de Wit J, Makino S, Yager N, et al. The autoimmune disease risk allele rs6897932 modulates monocyte IL7R surface and soluble receptor levels in a context-specific manner. bioRxiv 2018:262410.
- 27. Stürner KH, Borgmeyer U, Schulze C, Pless O, Martin R. A multiple sclerosis-associated variant of CBLB links genetic risk with type I IFN function. J Immunol 2014 Nov;193(9):4439-4447.
- 28. Alcina A, Abad-Grau MdelM, Fedetz M, Izquierdo G, Lucas M, Fernández O, et al. Multiple sclerosis risk variant HLA-DRB1*1501 associates with high expression of DRB1 gene in different human populations. PLoS One 2012;7(1):e29819.
- 29. Stankovic A, Dincic E, Ristic S, Lovrecic L, Starcevic Cizmarevic N, Djuric T, et al. Interleukin 7 receptor alpha polymorphism rs6897932 and susceptibility to multiple sclerosis in the Western Balkans. Multiple Sclerosis Journal 2010;16(5):533-536.
- 30. Al Jumah M, Al Balwi M, Hussein M, Kojan S, Al Khathaami A, Al Fawaz M, et al. Association of SNPs rs6498169 and rs10984447 with multiple sclerosis in Saudi patients: a model of the usefulness of familial aggregates in identifying genetic linkage in a multifactorial disease. Multiple Sclerosis Journal 2012;18(10):1395-1400.
- 31. Sanna S, Pitzalis M, Zoledziewska M, Zara I, Sidore C, Murru R, et al. Variants within the immunoregulatory CBLB gene are associated with multiple sclerosis. Nat Genet 2010 Jun;42(6):495-497.
- 32. Albor C, du Sautoy T, Kali Vanan N, Turner BP, Boomla K, Schmierer K. Ethnicity and prevalence of multiple sclerosis in east London. Multiple Sclerosis Journal 2017;23(1):36-42.
- 33. Wade BJ. Spatial analysis of global prevalence of multiple sclerosis suggests need for an updated prevalence scale. Mult Scler Int 2014;2014:124578.
- 34. Schiffer RB, Weitkamp LR, Wineman NM, Guttormsen S. Multiple sclerosis and affective disorder. Family history, sex, and HLA-DR antigens. Arch Neurol 1988 Dec;45(12):1345-1348.