Combined antiretroviral therapy (ART) is effective in controlling the progression of HIV disease and prolonging survival, but can be compromised by the development of drug resistance.1 A person can initially be infected with a drug-resistant HIV (primary resistance) or develop drug-resistant HIV after starting HIV medications (acquired resistance). Drug resistance occurs due to mutations in the genetic material of the virus. Just one mutation can make HIV resistant to some drugs like lamivudine and the non-nucleoside reverse transcriptase inhibitors (NNRTIs). However, HIV must go through a series of mutations to develop resistance to other drugs, including most protease inhibitors (PIs).2
Resistance to a drug diminishes the efficacy of that drug and often of members of the same drug class as well, thus limiting the options for constructing an effective subsequent treatment regimen. By reducing the efficacy of ART, morbidity, and mortality related to HIV infection increase and eventually, the risk for transmissibility also increase.3 Furthermore, the spread of resistance in the community negatively impacts the healthcare system as the need for expertise, and the cost of the drugs will increase. Some authors suggested testing all HIV patients with a viral load (VL) > 50 copies/mL for resistance to prevent such implications.4
HIV drug-resistance testing is recommended for persons with HIV infection at entry into care to guide the selection of the initial ART regimen.5,6 It is also recommended to guide therapy in patients with a suboptimal virologic response or virologic failure while on ART.5,6 Other common reasons for treatment failure other than drug resistance are attributed to non-adherence, drug absorption rates, drug activation, the patient’s metabolic rate, and interactions with other drugs.7
Genotypic resistance (GTR) tests are not available in all settings due to cost limitations. However, in recent years, they have become more common, faster, and cheaper. GTR testing was introduced for detection of both transmitted and acquired resistance at the Central Public Health Laboratories (CPHL) in Oman in September 2016. Before that, samples were sent abroad for resistance testing (primarily for acquired resistance).
There are many studies describing the prevalence of HIV drug resistance worldwide. A cohort study from the US concluded that among viremic patients an estimated 76% had resistance to one or more antiretroviral drugs.8 Locally, one study from Oman in 2004 included 29 HIV patients who were on ART for more than six months demonstrated a high rate of treatment failure contributed to factors other than resistance to ART such as non-adherence to therapy and treatment interruptions.9 Our current study is on a bigger sample of HIV treatment-experienced patients. The primary objective was to estimate the prevalence of HIV drug resistance and describe the common HIV genetic mutations in patients failing ART. We also investigated the impact of HIV resistance tests results on patients’ management.
Methods
This is a case record review limited to HIV drug resistance tests requested from a tertiary HIV center in Oman for patients failing ART. Treatment failure is defined as having an HIV VL more than 200 copies/mL after six or more months of ART initiation or modification.10
All HIV drug resistance tests requested between 1 April 2011 to 31 May 2017 were reviewed. Tests that were requested at baseline before starting ART were excluded because the aim of this study focused on the treatment failure, not the transmitted resistant strains. Baseline resistance testing was introduced recently to practice in Oman, and the small number of baseline resistance tests would not be enough for reliable estimation of ART resistance at the time of HIV diagnosis. In addition, inappropriate tests that were requested or repeated within six months of starting/switching therapy and tests that were reported as invalid results due to the VL being lower than the assay limit of detection (or other technical reasons) were also excluded. All other tests were included in the analysis.
Treatment failure was categorized as either failure to suppress (VL > 200 copies/mL six months after ART initiation/switch onwards) or viral rebound (two consecutive VL samples of > 200 copies/mL after achieving an initially undetectable VL six month after treatment initiation/switch). Data on timing from treatment failure to testing request were collected. HIV VL measurement and ART at the time of the test request were also collected. The latter was categorized based on the presence of thymidine analog (TA), zidovudine and/or stavudine, in the nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs) backbone. Patients who stopped their ART for more than four weeks were labeled as off ART.
Table 1: Patient characteristics and clinical profiles.
|
Sex (n = 84) |
|
|
|
Male |
63 |
75.0 |
|
Female |
21 |
25.0 |
|
Age, median (IQR), years |
38.0 (16.0) |
|
|
Risk factors for HIV (n = 84) |
|
Heterosexual |
38 |
45.2 |
|
Homo/bisexual |
10 |
11.9 |
|
Blood transfusion |
4 |
4.8 |
|
IVDU |
10 |
11.9 |
|
Multiple (sexual + IVDU) |
3 |
3.6 |
|
Vertical |
4 |
4.8 |
|
Not documented |
15 |
17.8 |
|
HBVsAg status (n = 84) |
|
Reactive |
2 |
2.4 |
|
Non-reactive |
79 |
94.0 |
|
Not documented |
3 |
3.6 |
|
HCV Ab status (n = 84) |
|
Reactive |
12 |
14.3 |
|
Non-reactive |
69 |
82.1 |
|
Not documented |
3 |
3.6 |
|
Baseline CD4, cells/μL (n = 84) |
|
< 200 |
43 |
51.2 |
|
200–349 |
18 |
21.4 |
|
350–499 |
10 |
11.9 |
|
> 500 |
10 |
11.9 |
|
Not available |
3 |
3.6 |
|
Baseline CD4, median (IQR), cells/μL |
195.0;
(103.0–348.0) |
|
|
Baseline VL, median (IQR), log |
4.8 (4.0–5.1) |
|
|
VL at test time, median (IQR), log |
4.8 (4.0–5.1) |
|
|
Type of treatment failure (n = 98) |
|
Failure to suppress |
70 |
71.4 |
|
Viral rebound |
28 |
28.6 |
|
Time from suppression failure till testing, median (IQR), months |
7.0
(2.0–29.0) |
|
|
Time from viral rebound till testing, median (IQR), months |
15.0
(5.0–43.0) |
|
|
Adherence at time of resistance test request (n = 98) |
|
Excellent |
1 |
1.0 |
|
Acceptable |
8 |
8.2 |
|
Poor |
71 |
72.4 |
|
Not documented |
18 |
18.4 |
|
ART at time of test request (n = 98) |
|
NNRTI + TA |
28 |
28.6 |
|
NNRTI + non-TA |
26 |
26.5 |
|
PI + TA |
4 |
4.1 |
|
PI + non-TA |
20 |
20.4 |
*Unless stated otherwise.
ART: antiretroviral therapy; IQR: interquartile range; IVDU: intravenous drug use; HIV: human immunodeficiency virus; HCV Ab: hepatitis C virus antibody; HBsAg: hepatitis B virus surface antigen; NRTI: nucleoside reverse transcriptase inhibitor; NNRTI: non-nucleoside reverse transcriptase inhibitor; PI: protease inhibitor; TA: thymidine analog; VL: viral load.
The data on GTR tests requested was collected from the hospital electronic patient record system (Alshifa). All GTR tests requested in the study period were reviewed. Tests meeting the inclusion criteria were further explored, and details related to those patients were obtained from the electronic patients’ record and the HIV clinic database. Variables collected include patient's demographics, risk factors for HIV (heterosexual, homo/bisexual), vertical transmission, intravenous drug use (IVDU), multiple factors (sexual and IVDU), blood transfusion, hepatitis B virus (HBV) and hepatitis C virus (HCV) status at diagnosis (positive HBV was defined as reactive hepatitis B surface antigen (HBsAg) and positive HCV was defined as reactive HCV antibodies), baseline CD4 count and VL (both defined as the measurement closest to the time of HIV diagnosis), and ART history and adherence to ART (adherence was defined as excellent, good, and poor if it was documented that patient forgot zero, 1–2, and > 2 doses per month, respectively). Data regarding the impact of the HIV drug resistance were collected. These data included the ART change (switch) timing and virological response (defined as VL < 200 copies/mL) six months after ART switch.
For the samples requested before September 2016, GTR testing was done at the Laboratoire Cerba in France (www.lab-cerba.com) using an in-house Sanger sequencing methodology according to the guidelines of the French National Agency for AIDS Research (ANRS). The interpretation was made according to the last version of the ANRS algorithm. All details of the protocol and the ANRS interpretation algorithm are available on the website (http://www.hivfrenchresistance.org/index.html).
Starting September 2016, the genotypic resistance test was performed at CPHL using a commercial assay (Applied Biosystems ViroSeq HIV-1 Genotyping System, Version 2.0) according to manufacturer’s instruction. After the initial step of reverse transcription and amplification, a total of seven primers covering the HIV-1 PI and reverse transcriptase areas were used for cycle sequencing incorporating BigDye sequencing reagents in the ABI PRISM 377. Assembly was done using the ViroSeq software and aligned by DNASTAR Lasergene. The final sequences were loaded in the Stanford HIV database for resistance interpretation. Because the segment of the HIV-1 genome used for genotypic testing (about 1500–2000 base pairs [bp]) is larger than that used for quantitative assays (about 100 bp), the sensitivity of most genotypic assays is lower (between 100 and 2000 RNA copies/mL depending on the assay).
The results of the resistance tests were classified as resistance to one class (either NRTIs or NNRTIs), two classes (NRTIs + NNRTIs), three classes (NRTIs + NNRTIs + PIs), or wild type (i.e., no resistance mutation was detected).
Initially, the data were anonymized and collected in hard copies, and then transcribed into the data collection software Epidata. The worksheet was rechecked against the hard copies for transcript inconsistencies. The frequencies and percentages were calculated using SPSS Statistics (IBM Corp. Released 2012. IBM SPSS Statistics for Windows, Version 21.0. Armonk, NY: IBM Corp).
Ethical approval was obtained from the hospital ethical committee. Written consent was unnecessary as this was a retrospective study and the tests were performed as the standard of care.
Results
Two-hundred and eighteen HIV drug resistance tests were requested from April 2011 to May 2017. One-hundred and twenty tests were excluded according to the exclusion criteria: 39 baseline tests, 32 failed amplification, and 49 other (e.g., inappropriate requests or technical reasons). A total of 98 tests were included in the analysis.
Table 2: Details of resistance tests results and patient follow-up.
|
NRTI only |
3 |
3.1 |
|
NNRTI only |
25 |
25.5 |
|
NRTI + NNRTI |
46 |
46.9 |
|
NRTI + NNRTI + PI |
8 |
8.2 |
|
Wild type |
16 |
16.3 |
|
ART switched (n = 82) |
|
|
|
Yes |
64 |
78.0 |
|
No |
18 |
22.0 |
|
Why no switch? (n = 18) |
|
|
|
No resistance to the current ART |
13 |
72.2 |
|
Not ready to start ART |
1 |
5.6 |
|
Lost to follow-up |
1 |
5.6 |
|
Only M184V |
1 |
5.6 |
|
Unknown reason |
2 |
11.1 |
|
Median time to switch, median (IQR), days |
29.5
(0.0–91.0) |
|
|
VL six months after switch,
copies/mL (n = 64) |
|
|
|
> 1000 |
17 |
26.6 |
|
200–1000 |
4 |
6.2 |
|
20–199 |
10 |
15.6 |
|
< 20 |
29 |
45.3 |
*Unless stated otherwise.
ART: antiretroviral therapy; IQR: interquartile range; NRTI: nucleoside reverse transcriptase inhibitor; NNRTI: non-nucleoside reverse transcriptase inhibitor; PI: protease inhibitor; VL: viral load.
Table 3: The genotype results.
|
A or A-like |
11 |
23.9 |
|
B or B-like |
6 |
13.0 |
|
C or C-like |
14 |
30.4 |
|
D or D-like |
5 |
10.9 |
|
G or G-like |
2 |
4.3 |
The characteristics of the 84 patients and the treatment and compliance details at the time of the 98 resistance tests included in this study are summarized in Table 1. Seventy-five percent of those patients were males and the median age was 38 years. Only two patients (2.4%) had HIV/HBV co-infection, while 12 (14.3%) had HIV/HCV co-infection. More than half of the patients (51.9%) started ART with a baseline CD4 of < 200 cells/µL, and the median baseline VL was log 4.8 copies/mL. The treatment failure was mainly due to initial failure of suppression (71.4%), and the majority of ART failure cases were non-adherent to medications (72.4%).
A total of 82 tests for 69 patients (83.7%; 95% CI: 76.4–91.0) showed resistance to at least one ART drug. Wild type was detected in only 16 cases; nine were off treatment at the time of the test. The impact of the resistance test on those patients’ management was followed. ART switch was done for the majority (78.0%) of cases with resistance virus (64 out of the 82 resistance cases). The median time from date of test request to date of ART switch was 29.5 days. VL was followed six months after ART switch and 39 out of 64 (60.9%) achieved virological response with a VL < 200 copies/mL [Table 2].
The genotype results were available for 46 patients only. The most prevalent subtype was subtype C or C-like (30.4%) followed by subtype A or A-like (23.9%). The details of the genotype results are summarized in Table 3.
The resistance tests listed the mutations responsible for the resistance to each class of ART. Since some patients had more than one resistance tests, the mutations are presented here as the number of patients who carry each mutation [Figure 1]. The most common mutations were M184V/I (45; 53.6%), K103N/S (41; 48.8%), and G190A/S/E (20; 23.8%).
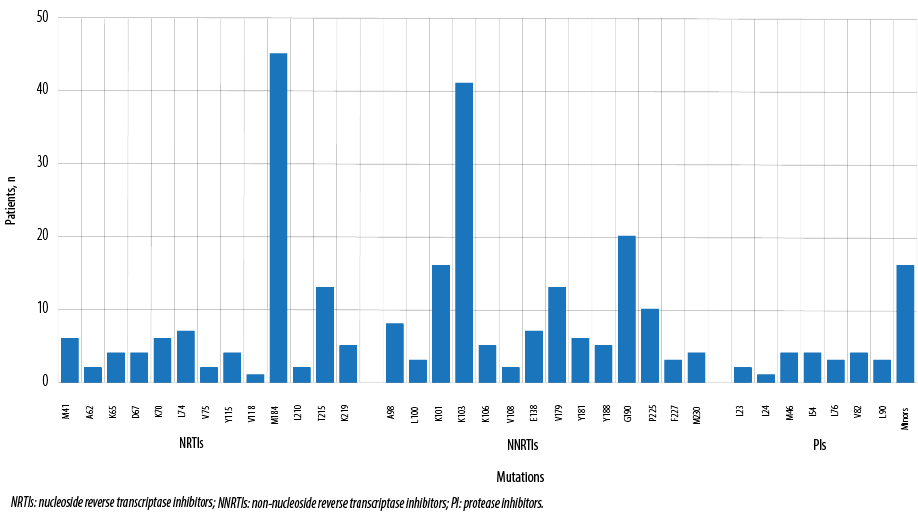
Figure 1: Mutations detected in patients with resistance to antiretroviral therapy.
Discussion
HIV drug resistance was common among patients failing ART and substitutions as residues 184, 103, and 190 were the most common resistant markers. Non-B subtypes were identified in 87.0% of the studied population. Most of our patients were diagnosed at late stages, evidenced by high VLs and low CD4 counts at baseline. Hence, the initiation of HIV treatment at earlier stages was not possible. The current HIV guidelines recommend early initiation of ART regardless of CD4 count.11
Non-adherence to medications played a major role in treatment failure and emergence of HIV resistance. The majority of those patients failing treatment were poorly adherent to the ART, and few of them claimed good adherence to the medications. It is difficult to estimate if acquired primary HIV resistance has contributed to treatment failure in this study due to lack of pre-ART HIV resistance test in most cases. The resistance tests were not considered immediately in most of those patients, but more in patients with viral rebound than those who failed to suppress within six months of treatment initiation. That could be explained in part by the fact that patients with viral rebound were likely to be given advice on adherence with close VL monitoring before resistance test request. This approach of continuation of failing regimens has led to the accumulation of more mutations.
This study highlights that HIV drug resistance should be suspected earlier because it is very common among HIV patients failing therapy. HIV resistance to at least one drug was detected in 83.7% of the tests requested during treatment failure. This is a higher prevalence compared to the 76.1% rate reported in a cohort study from the US; all were on ART at the time of testing.8 The rate identified in our study could be even higher because 56.3% of tests detected wild type viruses for patients who were off ART. It is possible that they are harboring archived mutations (i.e. those mutations are present but cannot be detected while the patient is off ART).
M184V/I, K103N/S, and G190A/S/E were the most common mutations detected among those patients. These mutations have been selected by the previous HIV drugs exposures. The most common HIV drugs used before the resistance tests were lamivudine, zidovudine, and efavirenz. A similar mutation pattern was described in another study from Brazil.12
ART was often changed according to the test result leading to good virological response (VL < 1000 copies/mL after six months) in 67% of cases. In the rest of the cases, VL remained > 1000 copies/mL due to poor adherence on the ART. This result points again to the issue of adherence as a major contributing factor leading to resistance and treatment failure. Patients’ education about drug resistance and the importance of adherence should be encouraged and maintained.
This study has limitations. First, it is a single-center study and may not reflect the prevalence of HIV drug resistance on treatment failure in Oman. To estimate the latter, more centers should be included with the proper sampling method. Another limitation was documentation bias and information unavailability, especially the details about the adherence to the ART. Thirdly, many patients had treatment failure, and their ART was switched without a resistance test result either because the tests were not requested or there were no valid results. Finally, the estimation of the exact impact of the drug resistance test on patient management might not be accurate because some patients defaulted from health care after the tests.
Conclusion
Our study highlights the importance of considering HIV drug resistance testing in patients failing ART. The main challenges are the early diagnosis of HIV, adherence to ART, and early response of HIV drug resistance. Educating patients about drug resistance may promote better adherence.
Disclosure
The authors declared no conflicts of interest. No funding was received for this study.
Acknowledgements
We thank Ms. Halima Al Sawafi, HIV clinic, Al Nahdha Hospital, for assistance with the data collection, CPHL staff for performing the HIV resistance testing, and Prof. Eskild Peterson, Department of Infectious Diseases, Royal Hospital, for comments that improved the manuscript.
references
- 1. Jensen-Fangel S, Pedersen L, Pedersen C, Larsen CS, Tauris P, Møller A, et al. Low mortality in HIV-infected patients starting highly active antiretroviral therapy: a comparison with the general population. AIDS 2004 Jan;18(1):89-97.
- 2. Drug resistance. IV/AIDS [Internet]. AIDSinfo 2018 [cited 2018 May 26]. Available from: https://aidsinfo.nih.gov/understanding-hiv-aids/fact-sheets/21/56/drug-resistance.
- 3. Wainberg MA, Friedland G. Public health implications of antiretroviral therapy and HIV drug resistance. JAMA 1998 Jun;279(24):1977-1983.
- 4. Assoumou L, Charpentier C, Recordon-Pinson P, Grudé M, Pallier C, Morand-Joubert L, et al. Prevalence of HIV-1 drug resistance in treated patients with viral load >50 copies/mL: a 2014 French nationwide study. J Antimicrob Chemother 2017;72(6):1769-1773.
- 5. EACS Guidelines [Internet]. 2018 [cited 2018 Jun 5]. Available from: http://www.eacsociety.org/guidelines/eacs-guidelines/eacs-guidelines.html.
- 6. Centers for Disease Control and Prevention (U.S.). Branson BM, Owen SM, Wesolowski LG, Bennett B, Werner BG, Wroblewski KE, et al. Laboratory testing for the diagnosis of HIV infection: updated recommendations [Internet]. Centers for Disease Control and Prevention. 2014 Jun [cited 2018 Jun 5]. Available from: http://stacks.cdc.gov/view/cdc/23447.
- 7. Schouten JT. HIV drug resistance and the other causes of treatment failure. STEP Perspect 1997;9(3):5-8.
- 8. Richman DD, Morton SC, Wrin T, Hellmann N, Berry S, Shapiro MF, et al. The prevalence of antiretroviral drug resistance in the United States. AIDS 2004 Jul;18(10):1393-1401.
- 9. Al Dhahry SH, Scrimgeour EM, Al Suwaid AR, Al Lawati MR, El Khatim HS, Al Kobaisi MF, et al. Human immunodeficiency virus type 1 infection in Oman: antiretroviral therapy and frequencies of drug resistance mutations. AIDS Res Hum Retroviruses 2004 Nov;20(11):1166-1172.
- 10. Guidelines for the use of antiretroviral agents in adults and adolescents living with HIV [Internet]. AIDSinfo 2018 [cited 2018 Aug 5]. Available from: https://aidsinfo.nih.gov/contentfiles/lvguidelines/adultandadolescentgl.pdf.
- 11. Initiation of antiretroviral therapy [Internet]. AIDSinfo 2017 [cited 2018 Oct 6]. Available from: https://aidsinfo.nih.gov/guidelines/html/1/adult-and-adolescent-arv/10/initiation-of-antiretroviral-therapy.
- 12. Cavalcanti AM, Lacerda HR, Brito AM, Pereira S, Medeiros D, Oliveira S. Antiretroviral resistance in individuals presenting therapeutic failure and subtypes of the human immunodeficiency virus type 1 in the Northeast Region of Brazil. Mem Inst Oswaldo Cruz 2007 Nov;102(7):785-792.