Diabetes encompasses a group of heterogeneous metabolic diseases characterized by chronic hyperglycemia. This hyperglycemia results from defects in insulin secretion, insulin action, or both. These defects are caused by a complex interplay of genetic and environmental factors linked to long-term damage, dysfunction, and failure of different organs, particularly the eyes, blood vessels, heart, nerves, and kidneys.1 The majority of those with diabetes have type 2 diabetes mellitus (T2DM).2 Usually, T2DM develops as a result of interactions of genetic and environmental risk factors, leading to relative deficiency of insulin with coexisting resistance to its activity.1 Additionally, certain ethnic groups have an increased risk of developing T2DM, which has been declared a global epidemic by the International Diabetes Federation. This multifaceted metabolic disorder stands out due to the inherent genetic predisposition within its population, a predisposition further exacerbated by the ongoing economic transition and rapid urbanization. Southeast Asia accommodates more than 90 million people with diabetes. Presently, Bangladesh ranks eighth in the list of countries with the highest prevalence of diabetes among adults aged 20–79, with 13.1 million documented cases. It is anticipated to ascend to seventh place by 2045.2,3
The etiology of T2DM is multifactorial, influenced by environmental, pathophysiological, and genetic factors. Genome-wide association studies have pinpointed numerous genes involved in T2DM, with the transcription factor 7-like 2 (TCF7L2) gene being one of the most susceptible.4 TCF7L2 stands out for its strong influence on T2DM risk and has been extensively studied in genetic linkage studies.5–9 Transcription factors are proteins that regulate gene expression by turning genes on or off. TCF7L2 is located on chromosome 10q25.3 and expressed in various organs, including fat cells, the liver, the gut, and pancreatic cells.10 TCF7L2 is a key player in the Wnt signaling pathway, which plays a crucial role in cell development and function.10 The exact mechanism by which TCF7L2 polymorphisms contribute to T2DM remains under investigation, but these variations are located in non-coding regions (introns) of the gene.11 TCF7L2 and GLP-1 are known to be involved in blood sugar control, and it is hypothesized that TCF7L2 mutations may affect insulin action, increasing T2DM susceptibility.12–14 Studies also suggested that TCF7L2 polymorphisms might influence pancreatic beta cells, leading to problems with glucose production and tolerance.15 The human TCF7L2 gene has at least four well-studied polymorphic markers associated with T2DM. Notably, among the well-studied TCF7L2 polymorphisms linked to T2DM are rs12255372 (G>T) in intron 4 and rs7903146 (C>T) in intron 3.7
Several studies have investigated the association of genetic variants, including TCF7L2, that could increase the risk of developing T2DM. The three single nucleotide polymorphisms (SNPs) of TCF7L2 (rs7903146 - intron 3, rs12255372, and rs11196205 - intron 4), individually and/or as a group, were found to be associated with T2DM in a wide spectrum of populations from different ethnicities: Finnish,5 Polish, Scandinavian, Chinese, Japanese,16 Swedish, Dutch, Palestinian, Iranian,6 Tunisian, Pakistani,9 and Indian populations.8,17
The link between T2DM and the SNP rs12255372, located in intron 4 of the TCF7L2 gene, was initially identified through a microsatellite study.18 Subsequently, this correlation has been corroborated in numerous studies conducted on diverse populations worldwide, as evidenced by previous studies.7,19 However, other studies did not detect any significant association in this context.20,21 Furthermore, a pharmacogenetic study suggested that individuals harboring the rs12255372 variant of TCF7L2 exhibited a suboptimal response to the widely prescribed oral anti-diabetic medication, sulfonylurea.22 These diverse outcomes underscore the imperative for further research into TCF7L2 variants among individuals from various ethnic backgrounds, including the population of Bangladesh.
To date, published data regarding the role of TCF7L2 SNPs in diabetes risk for the Bangladeshi population is scant. Therefore, this study aimed to determine the prevalence of TCF7L2 rs12255372 (G>T) in the adult Bangladeshi population with T2DM.
Methods
This cross-sectional study included 80 patients with T2DM and 80 normoglycemic controls without diabetes (the control group). The T2DM patients were recruited from the Department of Endocrinology outpatient facilities at a medical university. The control group was matched for age and sex with the case population. Diagnosis of T2DM was based on the standard criteria set by the American Diabetes Association (2021). The study protocol was approved by the Institutional Review Board of the medical university. Written informed consent was obtained from each participant before inclusion in the study. Patients diagnosed with type 1 diabetes or those with diabetes secondary to endocrinopathies, pancreatic diseases, or drug-induced diabetes were excluded from the study.
A comprehensive medical evaluation including age, sex, blood pressure, and anthropometric measurements (weight, height, and body mass index (BMI)) was performed on all T2DM patients and control participants. BMI was categorized as underweight (< 18.5 kg/m2), normal weight (18.5–22.9 kg/m2), overweight (23.0–27.4 kg/m2), and obese (≥ 27.5 kg/m2).23
Genomic DNA was extracted from peripheral blood leukocytes using the GeneJET Whole Blood Genomic DNA Purification Mini Kit (Thermo Fisher, USA). The amount of DNA was measured using a Qubit 2.0 fluorometer (Invitrogen, UK) and the Qubit dsDNA BR test kit 10. The rs12255372 polymorphism was genotyped using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). In brief, the targeted region was amplified using the following primers: Forward 5´- CTG GAA ACT AAG GCG TGA GG -3´; Reverse 5´- GGG TCG ATG TTG TTG AGC TT -3´. The PCR cycles consisted of five minutes at 94 °C, followed by 35 cycles of 30 seconds at 94 °C, 30 seconds at 54 °C, and 30 seconds at 72 °C. The final extension was performed at 72°C for 10 minutes. Subsequently, 10 μL of the 346 bp amplified product was digested with the Tsp509I restriction enzyme (Thermo Fisher Scientific, Lithuania) for three hours at 65 °C. Following enzyme treatment, the amplicons underwent 2% agarose gel electrophoresis and were stained with ethidium bromide. The studied region contains two restriction sites; the G>T allele change creates a new site. Some genotyping results were validated through direct sequencing. The expected product sizes after digestion were as follows: 143 bp and 104 bp for the normal homozygote (GG), 126 bp and 104 bp for the mutant homozygote (TT), and 143 bp, 126 bp, and 104 bp for the heterozygote (GT) [Figures 1 and 2]. Fragments < 100 bp were not visible. DNA extraction and molecular genotyping were performed at the Genetic Research Laboratory in the Department of Anatomy, Bangabandhu Sheikh Mujib Medical University, Bangladesh.
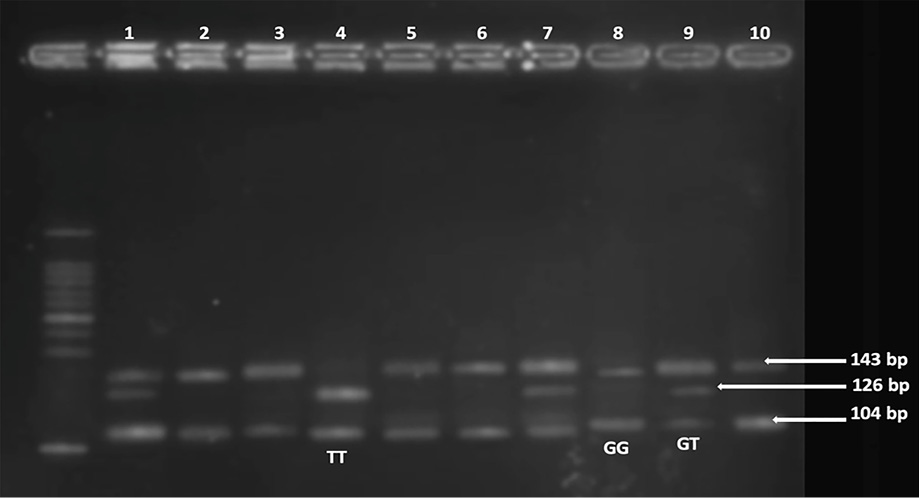 Figure 1: The polymerase chain reaction-restriction fragment length polymorphism analysis of transcription factor 7-like 2 rs12255372 (G>T) on a 2% agarose gel using the Tsp509I restriction enzyme in the type 2 diabetes mellitus group. In the gel electrophoresis results, bands at 143 bp and 104 bp in lanes 2,3,5,6,8, and 10 indicate the wild-type GG genotype. Lanes 1, 7, and 9 indicate the presence of the heterozygous GT genotype, with bands at 143 bp, 126 bp, and 104 bp. Lane 4 indicates TT, showing bands at 126 bp and 104 bp, corresponding to the homozygous (mutant) genotype.
Figure 1: The polymerase chain reaction-restriction fragment length polymorphism analysis of transcription factor 7-like 2 rs12255372 (G>T) on a 2% agarose gel using the Tsp509I restriction enzyme in the type 2 diabetes mellitus group. In the gel electrophoresis results, bands at 143 bp and 104 bp in lanes 2,3,5,6,8, and 10 indicate the wild-type GG genotype. Lanes 1, 7, and 9 indicate the presence of the heterozygous GT genotype, with bands at 143 bp, 126 bp, and 104 bp. Lane 4 indicates TT, showing bands at 126 bp and 104 bp, corresponding to the homozygous (mutant) genotype.
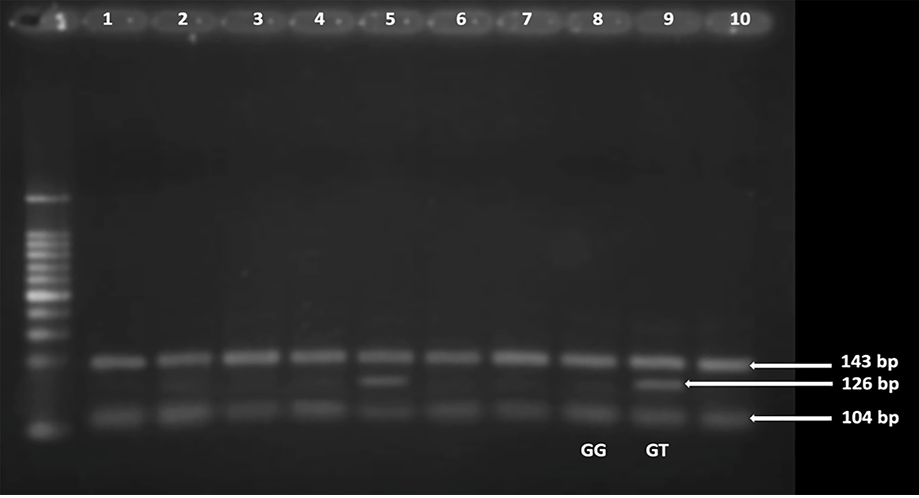 Figure 2: The polymerase chain reaction-restriction fragment length polymorphism analysis of transcription factor 7-like 2 rs12255372 in the control group. Lanes 1–4, 6–8, and 10 indicate the presence of the wild-type homozygous GG genotype. Lanes 5 and 9 show the presence of the heterozygote GT genotype.
Figure 2: The polymerase chain reaction-restriction fragment length polymorphism analysis of transcription factor 7-like 2 rs12255372 in the control group. Lanes 1–4, 6–8, and 10 indicate the presence of the wild-type homozygous GG genotype. Lanes 5 and 9 show the presence of the heterozygote GT genotype.
PCR-RFLP results were analyzed by gel electrophoresis to determine the frequency of the rs12255372 polymorphism. Qualitative values were expressed as frequencies and proportions, while quantitative values with a normal distribution were presented as mean and SD. Quantitative variables with a skewed distribution were described using median (IQR) values. The characteristics of cases and controls were compared using the chi-square (χ2) test of independence, independent sample Student’s t-test, and Mann–Whitney U test, as appropriate. The independent segregation of alleles was tested for Hardy-Weinberg equilibrium (HWE) using the chi-square goodness of fit test. The HWE assumes that a population is found to be in equilibrium when it is large and homogenous and does not experience notable mutation, migration, natural selection, or sexual selection, allowing genetic variations to remain constant across generations.24 The frequency of genotypes between the T2DM and control groups was compared using the chi-square test of independence within different genetic models. Statistical significance was determined by p-values < 0.05. All data were analyzed using SPSS (IBM Corp. Released 2019. IBM SPSS Statistics for Windows, Version 26.0. Armonk, NY: IBM Corp).
Results
This cross-sectional study included 80 patients with T2DM and 80 age- and sex-matched participants with normal glucose tolerance (controls) to assess the SNP rs12255372 (G>T) genotype frequencies and compare these between the two groups, including allele frequency and adherence to the HWE.
The median age of the studied participants was 44.0 years (IQR = 37.5–50.0), with 50.6% being female. There were no significant differences in age or sex between T2DM patients and controls. The minor T allele of the TCF7L2 rs12255372 variant significantly raised the risk of T2DM, with an allelic odds ratio (OR) of 3.29 (95% CI: 1.78–6.05;
p < 0.001). The T allele frequency was 28.1% in T2DM patients and 10.6% in controls.
Comparison of genotype frequencies in patients with T2DM versus controls revealed significant differences in the GT and TT genotypes, with OR = 3.00 (95% CI: 1.33–6.75; p = 0.007) and 5.26 (95% CI: 1.39–19.9; p = 0.008), respectively [Table 1].
Table 1: Genotype and allele frequency distribution of rs12255372 TCF7L2 (G>T) in T2DM cases and normoglycemic controls.
|
GG
|
46 (57.5)
|
66 (82.5)
|
Ref.
|
|
|
|
GT
|
23 (28.8)
|
11 (13.7)
|
3.00† (1.33–6.75)
|
7.39
|
0.007*
|
|
TT
|
11 (13.7)
|
3 (3.8)
|
5.26† (1.39–19.9)
|
7.06
|
0.008*
|
|
Major allele frequency (G)
|
115 (71.9)
|
143 (89.4)
|
Ref.
|
|
|
TCF7L2: transcription factor 7-like 2; G: guanine; T: thymine; T2DM: type 2 diabetes mellitus; OR: odds ratio.
*p-values were calculated using the chi-square test; †OR was calculated using the GG genotype as a reference.
To investigate the genetic model fitting the effect of TCF7L2 rs12255372, genotype frequencies were compared between the T2DM and control groups across different genetic models [Table 2]. Significant differences were observed between the T2DM and control groups across all four genetic models. Based on Akaike information criterion, the dominant genetic model was the best fit, as it yielded the lowest Akaike information criterion value. According to this model, the frequencies of variant homozygotes + heterozygotes (TT+GT) were 42.5% in the T2DM group and 17.5% in the control group, indicating the highest risk under the dominant model with OR = 3.48 (95% CI: 1.68–7.21; p < 0.001).
Table 2: Comparison of rs12255372 (G>T) genotype frequencies between the T2DM and control groups.
|
Co-dominant
|
|
|
|
|
|
|
|
G/G
|
112 (70.0)
|
46 (57.5)
|
66 (82.5)
|
1.00
|
0.001
|
215
|
|
G/T
|
34 (21.3)
|
23 (28.8)
|
11 (13.7)
|
3.00 (1.33–6.75)
|
|
|
|
T/T
|
14 (8.7)
|
11 (13.8)
|
3 (3.8)
|
5.26 (1.39–19.9)
|
|
|
|
Over-dominant
|
|
|
|
|
|
|
|
G/G + T/T
|
126 (78.7)
|
57 (71.2)
|
69 (86.3)
|
1.00
|
0.019
|
220.3
|
|
G/T
|
34 (21.3)
|
23 (28.8)
|
11 (13.7)
|
2.53 (1.13–5.63)
|
|
|
|
Dominant
|
|
|
|
|
|
|
|
G/G
|
112 (70.0)
|
46 (57.5)
|
66 (82.5)
|
1.00
|
< 0.001
|
213.6
|
|
G/T + T/T
|
48 (30.0)
|
34 (42.5)
|
14 (17.5)
|
3.48 (1.68–7.21)
|
|
|
|
Recessive
|
|
|
|
|
|
|
|
G/G + G/T
|
146 (91.3)
|
69 (86.3)
|
77 (96.2)
|
1.00
|
0.036
|
220.5
|
Comparison was done using chi-square (χ2) test of independence; ; G: guanine; T: thymine; T2DM: type 2 diabetes mellitus; OR: odds ratio AIC: Akaike information criterion.
Genotype frequencies violated the HWE in T2DM (χ2 = 6.67, df = 1; p = 0.010) and control subjects (χ2 = 6.09, df = 1; p = 0.010).
The clinical and biochemical features of T2DM patients and normoglycemic individuals were categorized based on rs12255372 (G>T) genotypes [Table 3]. Those having at least one variant type allele were found to have statistically significantly higher BMI, waist circumference and waist-hip ratio (WHR) compared to those having a pair of wild-type alleles (BMI of wild vs. mutant: 23.5 (21.6–25.7) vs. 26.3 (24.4–30.2); p < 0.001, WC of wild vs. mutant: 85.34 ± 9.00 vs. 96.52 ± 10.81; p < 0.001, and WHR of wild vs. mutant: 0.92 ± 0.05 vs. 0.97 ± 0.06; p < 0.001).
Table 3: Association of rs12255372 variants with clinical parameters in study participants.
|
BMI, kg/m2†
|
23.5 (21.6–25.7)
|
26.3 (24.4–30.2)
|
< 0.001
|
|
WC, cm#
|
85.34 ± 9.00
|
96.52 ± 10.81
|
< 0.001
|
|
WHR#
|
0.92 ± 0.05
|
0.97 ± 0.06
|
< 0.001
|
|
SBP, mmHg†
|
120 (110–125)
|
120 (120–125)
|
0.980
|
|
DBP, mmHg†
|
80 (70–80)
|
80 (75–85)
|
0.054
|
#Expressed in mean ± SD and comparison was done using independent sample Student’s t-test; †Data were expressed as median (IQR) and p-values were obtained using Mann-Whitney U test. G: guanine; T: thymine; BMI: body mass index; WC: waist circumference; WHR: waist-hip ratio; SBP: systolic blood pressure;
DBP: diastolic blood pressure; MBP: mean blood pressure.
Discussion
T2DM and its associated compilations are regarded as significant health challenges of the 21st century.25 Evidence from numerous studies strongly supports the notion that susceptibility to T2DM is heavily influenced by genetic factors.26 Though there is a high prevalence of T2DM in Bangladesh, limited epidemiological data on genetic predisposing factors of the disease are available.27 Of all the identified variants, TCF7L2 genetic variants have shown the most significant impact on the risk of T2DM, as reported in numerous studies.28–30 The present study explored the relationship between the rs12255372 variant in the TCF7L2 gene and T2DM within the Bangladeshi population.
Among T2DM participants, the observed genotype frequencies of wild-type rs12255372 (GG), heterozygous (GT), and homozygous variant (TT) were 57.5%, 28.8%, and 13.8%, respectively. Among the control group, the observed genotype frequencies of wild-type rs12255372, heterozygous, and homozygous variants were 82.5%, 13.7%, and 3.8%, respectively. In this polymorphism, G is the major allele and T is the minor allele. The association between rs12255372 and T2DM results of our study successfully replicate previous findings.6,25,27 Our study revealed the mutant T allele of rs12255372 had a frequency of 28.1%, which was comparable to the previous two studies on the Bangladeshi population where the frequency was found to be 29% and 19%, respectively,25,27 as well as populations of other countries including Southeast Asia, such as the Pakistani population (29%),31 the population of Hyderabad, India (24%),8 the Nigerian population (26%),32 the Iranian Kurdish ethnic group 18%,33 and the natives of India 26%.34
The aforementioned T allele was found to be substantially associated with the incidence of T2DM, with an OR of 3.29 (95% CI: 1.78–6.05; p < 0.001). Additionally, our study highlighted that homozygotes carry a significantly higher risk compared to heterozygote carriers. Therefore, our study supported the findings of previous studies regarding the connection between risk alleles. Moreover, it conformed to the multiplicative model of inheritance, suggesting that the risk for homozygotes is higher compared to heterozygotes across all SNPs. When the genotypes were compared, the wild-type homozygous GG genotype was predominant in the control group, while the heterozygous GT genotype and mutant TT homozygous genotype were more frequent in T2DM patients, and the difference was significant (p < 0.05).
The dominant, recessive, and codominant models were used to assess the associated risk between rs12255372 (G>T) genotypes of TCF7L2 gene and T2DM. In all three models, the TT and GT genotypes were found to confer a significant risk of susceptibility to T2DM. However, the dominant model showed the most significant risk of T2DM with OR = 3.48 (95% CI: 1.68–7.21; p < 0.001). Overall, this study showed a strong association between the rs12255372 (G>T) polymorphism of TCF7L2 gene and T2DM, and the finding is consistent with previous reports in Bangladesh and South-Asia region, as well as other different ethnic and geographical populations across the world. However, a modest association was noted in West-Africa and among African Americans with an OR below 2,35,36 whereas no association was detected in studies conducted on the Egyptian,37 Venezuelan,38 Arabic, South-African Zulu,39 Tunisian,40 and Brazilian populations.41 The observed regional variation may be attributed to a combination of genetic and environmental factors. Different populations may have distinct genetic backgrounds that influence the impact of this genetic variant. Additionally, lifestyle factors, dietary habits, rapid urbanization, and socioeconomic conditions can interact with genetic predisposition, contributing to varying T2DM susceptibility across regions.
The observed genotype frequencies were compared with the expected counts to determine the HWE of the TCF7L2 rs12255372 polymorphisms. The chi-square revealed a statistically significant p-value. Therefore, the genotype distribution of this polymorphism deviated from the HWE. Interpreting PCR-RFLP data can sometimes be challenging; thus, we opted to exclude all doubtful genotypes from our analysis. This finding was also reported in previous studies.42 This observation may be due to the limited sample size in our study or ongoing evolutionary processes.24 A study by Dalhat et al,31 conducted on a Pakistani population, revealed genotype frequencies of rs12255372 that were not in HWE. Deviation from HWE in that study was confirmed by replicated genotyping using fresh reagents, which yielded the same outcome.31 Another study by Nanfa et al,42 on the Cameroonian population, was also not in harmony with HWE. After validating their genotype analysis by sequencing, the authors concluded that the deviation from HWE could be due to small sample size or this polymorphism might have accumulated over generations, leading to deviation from HWE. Therefore, it is imperative to replicate our study findings using a larger sample size and more sensitive genotyping techniques, such as direct sequencing. This is particularly important given the occasional challenges in interpreting PCR-RFLP data. Careful selection of controls is necessary to prevent population stratification from creating confounding. Nevertheless, the high OR and statistically significant findings in our study strongly suggested an association between rs12255372 TCF7L2 and T2DM within our population.
To determine whether there is any interaction between rs12255372 (G>T) genotypes and clinical characteristics, the association between genotypes and covariates in the study participants was studied. Our study found that participants carrying the GT + TT genotypes of the rs12255372 variation had a significantly higher BMI and WHR as compared with those with the GG genotype. This result aligns with previously reported findings in an Iranian Kurdish ethnic group.33 However, several studies have not demonstrated an association between TCF7L2 rs12255372 polymorphism and obesity in European and US populations.43,44 A possible explanation for this incongruity could be the difference in ethnic background or the effects of environmental factors. South Asian populations have experienced a shift towards Westernized diets, characterized by high consumption of processed foods and low in take of nutrient-rich foods. Additionally, sedentary lifestyles, urbanization, and socioeconomic disparities have contributed to the rise in these health issues.
As a preliminary study, one of the limitations was the small sample size. This limitation might have restricted the distribution of genotypes in the population. Another limitation was that the design of our study was cross-sectional. A case-control study would be better to assess these associations. However, all our participants were sourced from a public hospital offering services to individuals from diverse socioeconomic backgrounds. Hence, our subjects had a strong potential to be representative of the general population of Bangladesh.
Conclusion
Since there has been limited prior genetic exploration of the population in this region, particularly regarding complex genetic disorders such as T2DM, it is imperative to investigate the involvement of various candidate genes in the development of T2DM. We found the minor T allele frequency of rs12255372 (G>T) among T2DM and control participants was about one-fourth and one-tenth, respectively, indicating that the rs12255372 (G>T) polymorphism of the TCF7L2 gene may be associated with the T2DM risk in the studied Bangladeshi population. Further explorations should aim to uncover the diversity within the genetic predisposition of the population in this region, which is characterized by significant variations in geography, ethnicity, culture, and genetics.
Disclosure
The authors declared no conflicts of interest. No funding was received for this study.
Acknowledgments
All authors acknowledge their gratitude to Dr. Shahed Morshed, Dr. Mukul Raihan, Dr. Sarojit Das, Dr. CFM Manzurur Rahim, Dr. Kamrul Azad, and all the staff members of the Endocrinology Department and the Anatomy Department for their help and support. We also thank all study participants.
references
- 1. Małecki M, Skupień J. Problems in differential diagnosis of diabetes types. Pol Arch Med Wewn 2008;118(7-8):435-440.
- 2. Sun H, Saeedi P, Karuranga S, Pinkepank M, Ogurtsova K, Duncan BB, et al. IDF diabetes atlas: global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res Clin Pract 2022 Jan;183:109119.
- 3. Ogurtsova K, Guariguata L, Barengo NC, Ruiz PL, Sacre JW, Karuranga S, et al. IDF diabetes atlas: global estimates of undiagnosed diabetes in adults for 2021. Diabetes Res Clin Pract 2022 Jan;183:109118.
- 4. Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007 Feb;445(7130):881-885.
- 5. Scott LJ, Bonnycastle LL, Willer CJ, Sprau AG, Jackson AU, Narisu N, et al. Association of transcription factor 7-like 2 (TCF7L2) variants with type 2 diabetes in a Finnish sample. Diabetes 2006 Sep;55(9):2649-2653.
- 6. Alami FM, Ahmadi M, Bazrafshan H, Tabarraei A, Khosravi A, Tabatabaiefar MA, et al. Association of the TCF7L2 rs12255372 (G/T) variant with type 2 diabetes mellitus in an Iranian population. Genet Mol Biol 2012 Apr;35(2):413-417.
- 7. Tong Y, Lin Y, Zhang Y, Yang J, Zhang Y, Liu H, et al. Association between TCF7L2 gene polymorphisms and susceptibility to type 2 diabetes mellitus: a large Human Genome Epidemiology (HuGE) review and meta-analysis. BMC Med Genet 2009 Feb;10(1):15.
- 8. Uma Jyothi K, Jayaraj M, Subburaj KS, Prasad KJ, Kumuda I, Lakshmi V, et al. Association of TCF7L2 gene polymorphisms with T2DM in the population of Hyderabad, India. PLoS One 2013;8(4):e60212.
- 9. Hameed T, Khan Z, Imran M, Ali S, Albegali AA, Ullah MI, et al. Associations of transcription factor 7-Like 2 (TCF7L2) gene polymorphism in patients of type 2 diabetes mellitus from Khyber Pakhtunkhwa population of Pakistan. Afr Health Sci 2021 Mar;21(1):15-22.
- 10. Del Bosque-Plata L, Hernández-Cortés EP, Gragnoli C. The broad pathogenetic role of TCF7L2 in human diseases beyond type 2 diabetes. J Cell Physiol 2022 Jan;237(1):301-312.
- 11. Jin T, Liu L. The Wnt signaling pathway effector TCF7L2 and type 2 diabetes mellitus. Mol Endocrinol 2008 Nov;22(11):2383-2392.
- 12. Gloyn AL, Braun M, Rorsman P. Type 2 diabetes susceptibility gene TCF7L2 and its role in beta-cell function. Diabetes 2009 Apr;58(4):800-802.
- 13. Shu L, Matveyenko AV, Kerr-Conte J, Cho J-H, McIntosh CH, Maedler K. Decreased TCF7L2 protein levels in type 2 diabetes mellitus correlate with downregulation of GIP- and GLP-1 receptors and impaired beta-cell function. Hum Mol Genet 2009 Jul;18(13):2388-2399.
- 14. Schäfer SA, Tschritter O, Machicao F, Thamer C, Stefan N, Gallwitz B, et al. Impaired glucagon-like peptide-1-induced insulin secretion in carriers of transcription factor 7-like 2 (TCF7L2) gene polymorphisms. Diabetologia 2007 Dec;50(12):2443-2450.
- 15. Zhou Y, Park S-Y, Su J, Bailey K, Ottosson-Laakso E, Shcherbina L, et al. TCF7L2 is a master regulator of insulin production and processing. Hum Mol Genet 2014 Dec;23(24):6419-6431.
- 16. Kunika K, Tanahashi T, Numata S, Ueno SI, Ohmori T, Nakamura N, et al. Common coding variant in the TCF7L2 gene and study of the association with type 2 diabetes in Japanese subjects. J Hum Genet 2008;53(11-12):972-982.
- 17. Bodhini D, Radha V, Dhar M, Narayani N, Mohan V. The rs12255372(G/T) and rs7903146(C/T) polymorphisms of the TCF7L2 gene are associated with type 2 diabetes mellitus in Asian Indians. Metabolism 2007 Sep;56(9):1174-1178.
- 18. Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 2006;38(3):320-323.
- 19. Peng S, Zhu Y, Lü B, Xu F, Li X, Lai M. TCF7L2 gene polymorphisms and type 2 diabetes risk: a comprehensive and updated meta-analysis involving 121,174 subjects. Mutagenesis 2013 Jan;28(1):25-37.
- 20. Alsmadi O, Al-Rubeaan K, Mohamed G, Alkayal F, Al-Saud H, Al-Saud NA, et al. Weak or no association of TCF7L2 variants with type 2 diabetes risk in an Arab population. BMC Med Genet 2008 Jul;9:72.
- 21. Guo T, Hanson RL, Traurig M, Muller YL, Ma L, Mack J, et al. TCF7L2 is not a major susceptibility gene for type 2 diabetes in Pima Indians: analysis of 3,501 individuals. Diabetes 2007 Dec;56(12):3082-3088.
- 22. Pearson ER, Donnelly LA, Kimber C, Whitley A, Doney AS, McCarthy MI, et al. Variation in TCF7L2 influences therapeutic response to sulfonylureas: a GoDARTs study. Diabetes 2007 Aug;56(8):2178-2182.
- 23. Expert Consultation WH; WHO Expert Consultation. Appropriate body-mass index for Asian populations and its implications for policy and intervention strategies. Lancet 2004 Jan;363(9403):157-163.
- 24. Smith MU, Baldwin JT. Making sense of Hardy-Weinberg equilibrium. Am Biol Teach 2015;77(8):577-582.
- 25. Salauddin A, Chakma K, Hasan MM, Akter F, Chowdhury NA, Chowdhury SR, et al. Association between TCF7L2 polymorphism and type 2 diabetes mellitus susceptibility: a case-control study among the Bangladeshi population. Mol Biol Rep 2023 Jan;50(1):609-619.
- 26. Kaur J, Kaur J, Mittal M, Kaur N, Singh T. Type 2 diabetes mellitus and genetics: TCF7L2 a candidate gene. New Advances in Medicine and Medical Science 2023;3:143-149.
- 27. Barman N, Atiqul Haque M, Firoz M, Abdullah Yusuf M, Islam AB. Association of transcription factor 7-like 2 rs12255372 polymorphism with susceptibility of type 2 diabetes mellitus in Bangladeshi population. Mol Genet Genomics 2023 Sep;298(5):1201-1209.
- 28. Zheng X, Ren W, Zhang S, Liu J, Li S, Li J, et al. Association of type 2 diabetes susceptibility genes (TCF7L2, SLC30A8, PCSK1 and PCSK2) and proinsulin conversion in a Chinese population. Mol Biol Rep 2012 Jan;39(1):17-23.
- 29. Barra GB, Dutra LA, Watanabe SC, Costa PG, Cruz PS, Azevedo MF, et al. Association of the rs7903146 single nucleotide polymorphism at the transcription factor 7-like 2 (TCF7L2) locus with type 2 diabetes in Brazilian subjects. Arq Bras Endocrinol Metabol 2012 Nov;56(8):479-484.
- 30. Kaur N, Bhatti GK, Kaur S, Bhadada SK, Singh S, Bhatti JS. Transcription factor 7-like 2 gene, rs12255372 (G/T) variant and susceptibility to type 2 diabetes mellitus in North Indians. Gene Rep 2020;19:100595.
- 31. Dalhat M, Bashiru I, Bello H, Saidu Y, Abbas A. Association of transcription factor 7 like 2 (TCF7L2) rs12255372 (G/T) gene polymorphism and type 2 diabetes mellitus. Journal of Advances in Biology Biotechnology 2017 Jan;15(4):1-7.
- 32. Engwa G, Nwalo F, Ozokonkwo C, Agbafor K, Nkeh-Chungag B, Ubi B. Association of TCF7L2 rs12255372–G/T polymorphism with type 2 diabetes in a Nigerian population. Genet Mol Res 2021;20(1):1-10.
- 33. Shokouhi S, Delpisheh A, Haghani K, Mahdizadeh M, Bakhtiyari S. Association of rs7903146, rs12255372, and rs290487 polymorphisms in TCF7L2 gene with type 2 diabetes in an Iranian Kurdish ethnic group. Clin Lab 2014;60(8):1269-1276.
- 34. Bodhini D, Chidambaram M, Liju S, Prakash VG, Gayathri V, Shanthirani CS, et al. Association of TCF7L2 polymorphism with diabetic nephropathy in the South Indian population. Ann Hum Genet 2015 Sep;79(5):373-379.
- 35. Helgason A, Pálsson S, Thorleifsson G, Grant SF, Emilsson V, Gunnarsdottir S, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet 2007 Feb;39(2):218-225.
- 36. Sale MM, Smith SG, Mychaleckyj JC, Keene KL, Langefeld CD, Leak TS, et al. Variants of the transcription factor 7-like 2 (TCF7L2) gene are associated with type 2 diabetes in an African-American population enriched for nephropathy. Diabetes 2007 Oct;56(10):2638-2642.
- 37. Bawady SA, Mahmoud NH, Alzayet MH, Radwan RA, Abdel-Wahed MA. Relationship of transcription factor 7-like-2 (TCF7L2) gene polymorphism rs12255372 and glycemic control in type 2 diabetes mellitus. Egypt J Hosp Med 2022;88(1):2838-2844.
- 38. Moran Y, Labrador L, Camargo ME, Fernández D, Chiurillo MA. Design of an allele-specific PCR assay to genotype the rs12255372 SNP in a pilot study of association between common TCF7L2 polymorphisms and type 2 diabetes in Venezuelans. Arch Endocrinol Metab 2015 Jul;60(3):246-251.
- 39. Pirie F, Motala A, Pegoraro R, Paruk I, Govender T, Rom L. Variants in PPARG, KCNJ11, TCF7L2, FTO, and HHEX genes in South African subjects of Zulu descent with type 2 diabetes. Afr J Diabetes Med 2010;18(1):12-16.
- 40. Kifagi C, Makni K, Boudawara M, Mnif F, Hamza N, Abid M, et al. Association of genetic variations in TCF7L2, SLC30A8, HHEX, LOC387761, and EXT2 with type 2 diabetes mellitus in Tunisia. Genet Test Mol Biomarkers 2011 Jun;15(6):399-405.
- 41. Barros CM, Araujo-Neto AP, Lopes TR, Barros MA, Motta FJ, Canalle R, et al. Association of the rs7903146 and rs12255372 polymorphisms in the TCF7L2 gene with type 2 diabetes in a population from northeastern Brazil. Genet Mol Res 2014 Sep;13(3):7889-7898.
- 42. Nanfa D, Sobngwi E, Atogho-Tiedeu B, Noubiap JJ, Donfack OS, Mofo EP, et al. Association between the TCF7L2 rs12255372 (G/T) gene polymorphism and type 2 diabetes mellitus in a Cameroonian population: a pilot study. Clin Transl Med 2015 Apr;4(1):17.
- 43. Ngwa EN, Sobngwi E, Atogho-Tiedeu B, Noubiap JJ, Donfack OS, Guewo-Fokeng M, et al. Association between the rs12255372 variant of the TCF7L2 gene and obesity in a Cameroonian population. BMC Res Notes 2015 Nov;8(1):717.
- 44. Klünder-Klünder M, Mejía-Benitez MA, Flores-Huerta S, Burguete-García AI, García-Mena J, Cruz M. rs12255372 variant of TCF7L2 gene is protective for obesity in Mexican children. Arch Med Res 2011 Aug;42(6):495-501.