Chronic liver disease and cirrhosis remain major health concerns, consistently being among the top causes of death worldwide.1 Despite significant progress in medical treatments, the mortality rates associated with liver diseases have not shown significant improvement over the past three decades.2 To reduce mortality in patients with chronic liver disease and cirrhosis, liver transplantation is a potential solution.3,4 However, often the treating physicians have to weigh the risks and benefits of early liver transplantation in patients who present with evidence of physiological decompensation. This is essential because in some cases the time window between the intervention and the patient’s death may be limited. Failing to address these factors promptly may render intensive care unit (ICU) care and advanced interventions such as liver transplantation futile for some of these high-risk individuals.5,6 Therefore, it is necessary to establish a rational basis for discontinuing ICU care based on futility. Various scores, including Child–Turcotte–Pugh (CTP) and Mayo End-Stage Liver Disease (MELD), have been developed to assess prognosis and guide treatment prioritization including liver transplantation.7 Research suggests that the Chronic Liver Failure Consortium Acute-on-Chronic Liver Failure (CLIF-C ACLF) score might outperform both CTP and MELD in predicting short and medium-term mortalities in a range of patient scenarios.8,9 While these scores offer valuable information, they lack accuracy as they group patients with different levels of disease severity into similar risk categories. Also, there are no prognostic models that offer personalized estimates of the risk of liver-related death for individuals with alcohol-associated cirrhosis.10 In addition, the traditional prognostic scores may not take into account some of the risk factors associated with short-term mortality, such as acute kidney injury, plasma ammonia level, sarcopenia, and increased platelet aggregation.11,12
Advanced statistical tools and machine learning (ML) techniques are found to be superior to traditional statistical methods in predicting the risk of mortality. By capturing higher-dimensional and potentially nonlinear effects of variables, they can process more variables.13 Artificial intelligence (AI) is an expanding domain of information technology, widely used in various sectors such as e-commerce, media, and finance. Although health sciences have been slower to adopt AI, particularly ML, it is now gaining attention. ‘Traditional’ ML trains models through mathematical functions/rulesets that employ non-linear relationships, yielding more precise classification and prediction outputs compared to classical statistics.14 Deep learning, a relatively new subset of ML, employs deep neural networks to analyze data through multiple layers of interconnected artificial neurons, mimicking the structure and function of the human cerebral cortex.
A patient with cirrhosis represents a complex, multidimensional system for prognosis prediction. Emerging research suggests that AI and ML should be capable of providing superior mortality predictions for individual cirrhosis cases.15,16 Yet, previous studies have been limited in scope, which have used AI and ML focused on 90-day mortality prediction.5,15,17 There is a need for more comprehensive investigations using AI and ML to develop more accurate models capable of predicting short-term mortality risk in cirrhosis cases.17,18 Therefore, the objective of this study is to develop an AI-ML-based model to predict 28-day mortality in cirrhosis patients.
Methods
This retrospective study was conducted at Sultan Qaboos University Hospital (SQUH), a multispecialty teaching hospital in Oman.19,20 The subjects comprised all adult patients diagnosed with acute decompensation of liver cirrhosis and hospitalized in SQUH from January 2015 to December 2021. The study was approved by the Medical Research Ethics Committee of the College of Medicine and Health Sciences of Sultan Qaboos University (SQU EC/349/2021 MREC #2375).
The methodology described by Al Kaabi et al,2 in 2023 was adopted. In case of multiple hospitalizations, the first admission with acute decompensation was taken as the index admission. The required data was extracted from the patients’ electronic health records. This included demographic details, admission diagnosis, relevant comorbidities, cirrhosis etiology, duration of hospital stay, intensive care requirements, and relevant laboratory results.
The outcome of interest was 28-day all-cause mortality during the follow-up period. This was ascertained by reviewing the electronic health records of patients or through phone calls where necessary.
Before proceeding with model development, the dataset was refined. Missing values in binary features were imputed using with the mode, while continuous features had their missing values imputed using the median. To ensure uniformity in range, continuous features were scaled using the MinMaxScaler.
Four general ML algorithms, namely, Decision Tree, Random Forest, Logistic Regression, and Naïve Bayes, were employed to develop the models. These algorithms were chosen for their ability to handle both continuous and binary features and their capability to capture complex relationships within the data.21 In addition, a deep learning algorithm that incorporated multilayer artificial neural networks (ANNs) and recurrent neural network (RNN) including long short-term memory (LSTM) was implemented to test the ability of neural networks in capturing non-linear dependencies in the data.22 For each algorithm, the dataset was split into training and testing sets at 80:20 ratio.
Various metrics were calculated to compare the models' performance, including accuracy, precision, recall, and F1-Score. Accuracy measures the overall correctness of predictions while precision quantifies the proportion of correctly predicted positive cases. Recall measures the sensitivity of the model in identifying positive cases. F1-score combines precision and recall into a single metric. The metrics were calculated for each model and reported. To provide a visual representation of the performance, bar plots were generated for the four metrics with each bar representing the performance of a given model.22
Categorical variables were expressed in numerical values and percentages while continuous variables were presented as means for normally distributed data and medians with IQR for non-normally distributed data. To compare the continuous variables between the two groups, the student’s t-test was employed for normally distributed variables, and the Wilcoxon rank-sum test for non-normally distributed variables. The chi-square test assessed the relationship between categorical variables. Statistical significance was set at a two-sided p-value < 0.050. All statistical analyses were conducted using Stata (StataCorp. 2023. Stata Statistical Software: Release 18. College Station, TX: StataCorp LLC).
For the development of the models and their evaluation, we used Anaconda distribution, including Python programming language and various libraries. We initially developed our models using four general ML algorithms, namely Decision Tree, Random Forest, Naïve Bayes, and Logistic Regression. Multilayer ANNs, LSTM, and RNN were employed as deep learning algorithms. Each generated model was trained and tested using a dataset containing features related to acute decompensations of chronic liver disease.
Results
The subjects comprised of 173 patients admitted with acute decompensated liver disease during the study period. The demographic, clinical and laboratory findings, and factors associated with 28-day mortality are reported in Table 1.
Table 1: Cirrhosis patients’ characteristics, and relevant clinical and laboratory findings (N = 173).
|
General characteristics
|
|
Age, years (mean ± SD)
|
58.0 ± 13.8
|
62.0 ± 13.7
|
57.0 ± 13.7
|
0.050
|
|
Male patients
|
124 (71.7)
|
30 (83.3)
|
94 (68.6)
|
0.080
|
|
Mean weight, kg (range)
|
69.3 (84.0–60.0)
|
70 (60.0–81.65)
|
69.2 (60.8–85.0)
|
0.510
|
|
Mean body mass index (range)
|
27.7 (31.8–22.4)
|
28.1 (22.1–30.5)
|
27.5 (22.6–32.0)
|
0.440
|
|
Comorbidities
|
|
Hypertension
|
77 (44.5)
|
12 (33.3)
|
65 (47.4)
|
0.130
|
|
Diabetes mellitus
|
75 (43.4)
|
14 (38.9)
|
61 (44.5)
|
0.540
|
|
Ischemic heart disease
|
36 (20.8)
|
9 (25.0)
|
27 (19.7)
|
0.490
|
|
Chronic kidney disease
|
20 (11.6)
|
5 (13.9)
|
15 (10.9)
|
0.620
|
|
Smoking
|
30 (17.3)
|
10 (27.8)
|
20 (14.6)
|
0.060
|
|
Etiology of liver cirrhosis
|
|
Alcohol dependence
|
51 (29.5)
|
13 (36.1)
|
38 (27.7)
|
0.330
|
|
Hepatitis B virus
|
46 (26.6)
|
7 (19.4)
|
39 (28.5)
|
0.270
|
|
Hepatitis C virus
|
48 (27.7)
|
13 (36.1)
|
35 (25.5)
|
0.210
|
|
Non-alcoholic fatty liver disease
|
24 (13.9)
|
5 (13.9)
|
19 (13.9)
|
1.000
|
|
Reason for admission
|
|
Spontaneous bacterial peritonitis
|
15 (8.7 )
|
4 (11.1)
|
11 (8.0)
|
0.520
|
|
Hepatic encephalopathy
|
68 (39.3)
|
27 (75.0)
|
41 (29.9)
|
< 0.001
|
|
Ascites
|
111 (64.2)
|
26 (72.2)
|
85 (62.0)
|
0.260
|
|
Variceal bleeding
|
85 (49.1)
|
12 (33.3)
|
73 (53.3)
|
0.030
|
|
Course of admission
|
|
Intensive care unit
|
34 (19.7)
|
24 (66.7)
|
10 (7.3)
|
< 0.001
|
|
Mechanical ventilation
|
31 (17.9)
|
22 (61.1)
|
9 (6.6)
|
< 0.001
|
|
Length of hospital stay, days (range)
|
7.0 (4.0–12.0)
|
10.5 (6.5–23.5)
|
6.0 (4.0–11.0)
|
0.003
|
|
Treatments given
|
|
Beta-blockers
|
84 (48.6)
|
12 (33.3)
|
72 (52.6)
|
0.040
|
|
Diuretics
|
114 (65.9)
|
24 (66.7)
|
90 (66.0)
|
0.913
|
|
Lactulose
|
134 (77.5)
|
34 (94.4)
|
100 (73.0)
|
0.006
|
|
Clinical, hematological, and biochemical profile, mean (range)
|
|
FiO2
|
0 (0.0)
|
0.3 (0.0–1.0)
|
0 (0.0)
|
< 0.001
|
|
Mean arterial pressure
|
71.0 (67.0–82.0)
|
58.5 (47.0–71.5)
|
72.0 (69.0–84.0)
|
< 0.001
|
|
Hemoglobin, g/dL
|
10.2 ± 2.5
|
10.3 ± 2.5
|
10.2 ± 2.5
|
0.827
|
|
Platelets, 109/L
|
154.5 (211.5–99.5)
|
175.0 (112.0–235.0)
|
136.0 (93.0–205.0)
|
0.086
|
|
White cell count, 109/L
|
7.2 (10.7–5.1)
|
9.3 (7.1–15.5)
|
6.8 (5.0–9.1)
|
< 0.001
|
|
International normalised ratio
|
1.3 (1.5–1.2)
|
1.5 (1.3–1.9)
|
1.3 (1.2–1.5)
|
0.001
|
|
Prothrombin time, seconds
|
14.2 (12.6–16.5)
|
15.6 (14.4–20.4)
|
14.0 (12.5–15.6)
|
0.001
|
|
Creatinine, mmol/L
|
72.0 (106.0–57.0)
|
82.5 (58.0–119.5)
|
71.0 (57.0–99.0)
|
0.190
|
|
Sodium, mmol/L
|
135.0 (131.0–138.0)
|
132.5 (128.5–137.0)
|
135.0 (132.0–138.0)
|
0.151
|
|
Potassium, mmol/L
|
4.3 (3.9–4.8)
|
4.7 (3.9–5.1)
|
4.3 (3.9–4.7)
|
0.101
|
|
Alanine aminotransferase, IU/L
|
37.0 (24.0–70.0)
|
51.0 (34.0–109.0)
|
32.5 (24.0–61.0)
|
0.005
|
|
Albumin, g/L
|
30 (25–34)
|
25 (22–31)
|
31 (25–35)
|
< 0.001
|
|
Alkaline phosphatase, IU/L
|
136 (100–220)
|
220 (129–392)
|
125 (96–180)
|
0.001
|
|
Aspartate aminotransferase, IU/L
|
63 (42–134)
|
110 (70–232)
|
55 (39–118)
|
< 0.001
|
|
Bilirubin, umol/L
|
34 (17–79)
|
49 (22–180)
|
29 (15–73)
|
0.018
|
|
Gamma-glutamyl transferase, IU/L
|
230 (75–481)
|
258 (68–809)
|
215 (75–465)
|
0.603
|
|
HbA1c, % (range)
|
6.4 (5.0–8.4)
|
5.7 (4.7–7.2)
|
6.4 (5.2–8.4)
|
0.500
|
|
Liver cirrhosis scores
|
|
CTP, score (range)
|
9 (7–11)
|
10 (9–12)
|
8 (7–10)
|
< 0.001
|
|
MELD-Na, score (range)
|
18 (13–25)
|
24 (18–29)
|
17 (12–24)
|
0.001
|
FiO2: fraction of inspired oxygen; HbA1c: glycated hemoglobin CTP: child-turcotte-pugh; MELD-Na: model for end-stage liver disease-sodium; CLIF-C: chronic liver failure consortium.
The scores of various ML models under different assessment scales are listed in Table 2. The Decision Tree model achieved an accuracy of 71.4%, with a precision of 38.5% and a recall of 71.4%. The F1-score for this model was 0.500. The Logistic Regression model showed higher performance with an accuracy of 82.9%. It achieved a precision of 55.6% and a recall of 71.4%. The F1-score was 0.625. Both the Naïve Bayes and Random Forest models performed similarly, both achieving an accuracy of 82.9% and a precision of 54.5%. The Naïve Bayes model showed a higher recall of 85.7%, resulting in an F1-score of 0.667. The Random Forest model had a recall of 85.7% and an F1-score of 0.667. The multilayer ANNs model achieved an accuracy of 74.3%, with a precision of 37.5% and a recall of 42.9%. The F1-score was 0.400. In contrast, the RNN and LSTM models exhibited lower performance metrics. The RNN model achieved an accuracy of 80.0% with a perfect precision of 100%. However, it had a recall of 0.0% and an F1-score of 0.000. The LSTM model also had an accuracy of 80.0% but performed poorly in terms of precision, recall, and the F1-score, all of which were 0.000.
Table 2: Comparison of performance of various machine learning and deep learning models in predicting 28-day mortality.
|
Decision Tree
|
0.714
|
0.385
|
0.714
|
0.500
|
0.714
|
|
Logistic Regression
|
0.829*
|
0.556*
|
0.714
|
0.625
|
0.786
|
|
Naïve Bayes
|
0.829*
|
0.545
|
0.857*
|
0.667*
|
0.839*
|
|
Random Forest
|
0.829*
|
0.545
|
0.857*
|
0.667*
|
0.839*
|
|
Multilayer ANN
|
0.743
|
0.375
|
0.429
|
0.400
|
0.625
|
|
RNN
|
0.800
|
1.000
|
0.000
|
0.000
|
0.648
|
ANN: artificial neural network; RNN: recurrent neural network; LSTM: long short-term memory; AUROC: area under the receiver operating characteristics curve; *: top score.
Among the traditional ML models (those without Deep Learning), the Logistic Regression, Naïve Bayes, and Random Forest showed high AUROC values indicating good discrimination capability in predicting mortality. In contrast, the deep learning models— multilayer ANNs, RNN, and LSTM — demonstrated a lower capacity for discrimination, as revealed by their lower area under the receiver operating characteristics curve values [Table 2]. Overall, Naïve Bayes and Random Forest showed the highest performance metrics among the traditional ML models.
The Feature Importance Analysis using the Random Forest model revealed the top 10 contributing features for mortality prediction in patients with chronic liver disease and cirrhosis following admission with acute decompensation. The most influential feature was ICU admission (importance: 0.112), followed by mechanical ventilation (importance: 0.095), and mean arterial pressure (importance: 0.073). Other significant features included platelet count (importance: 0.063), alkaline phosphatase (importance: 0.056), bilirubin (importance: 0.054), and white blood cell count (importance: 0.048). Figure 1 shows the top 10 contributing features in the order of relative importance.
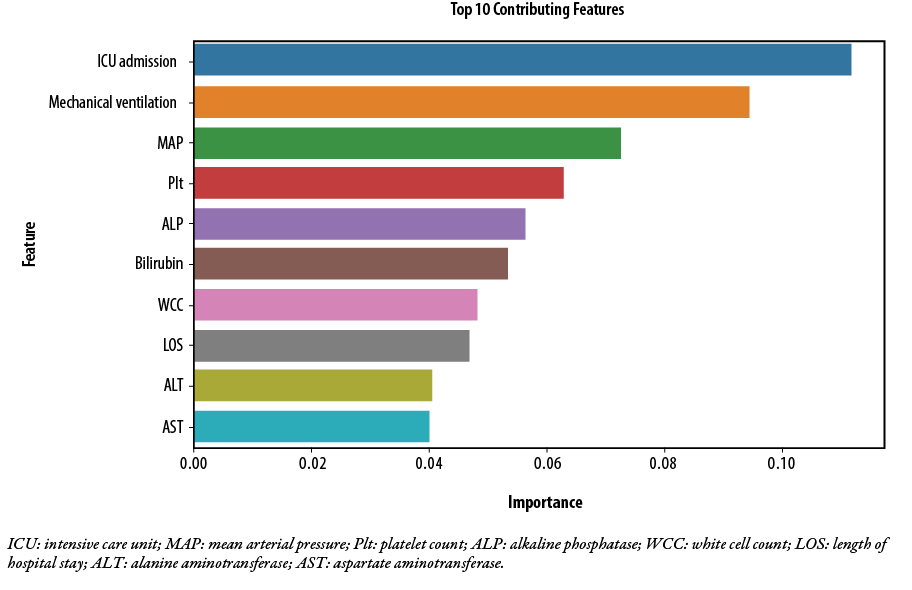 Figure 1: Top 10 predictors of 28 days mortality for hospitalized patients with acute decompensation of liver cirrhosis, as revealed by Feature Importance Analysis using Random Forest model.
Figure 1: Top 10 predictors of 28 days mortality for hospitalized patients with acute decompensation of liver cirrhosis, as revealed by Feature Importance Analysis using Random Forest model.
Discussion
Our study used traditional ML as well as deep learning techniques to develop a model to predict 28-day mortality of hospitalized patients with acute decompensation of liver cirrhosis. We evaluated the performance of various models, including Decision Tree, Logistic Regression, Naïve Bayes, Random Forest, multilayer ANNs, RNN, and LSTM, using a dataset that included demographic, clinical, and laboratory features related to chronic liver disease.
The Naïve Bayes and Random Forest models had the highest performance metrics among the traditional ML models. They achieved high accuracy, precision, recall, and F1-score. Despite its simplicity, the Naïve Bayes model was able to capture relevant patterns in the data and make accurate predictions for mortality.
The AUROC values further confirmed the discriminatory ability of these models with values ranging from 0.786 to 0.839. The Naïve Bayes model, although simpler in its assumption of feature independence, delivered competitive results. It showed a balanced trade-off between precision and recall, similar to the Decision Tree model. It is worth noting that the AUROC values for predicting 28-day mortality using the Naïve Bayes and Random Forest models were comparable to the AUROC values for CLIF-C and superior to those for CTP and MELD-Na within the same cohort of patients, as previously reported.2
In a 2021 study on a cohort of 2170 cirrhosis patients,23 the prediction of 90-day mortality was examined through the utilization of three AI models: Logistic Regression, Kernel Support Vector Machine, and Random Forest Classifiers. The study yielded a modest AUROC score of 0.67 for the prediction of 90-day mortality. The mortality predictions by the AI models were only as accurate as those by MELD-Na, a popular non-AI model.
The deep learning models used in our study—including the multilayer ANNs, RNN, and LSTM—showed lower performance metrics. The multilayer ANNs model achieved moderate performance, while the RNN and LSTM models showed poor performance, with recall and F1-score of 0 for predicting mortality. Meanwhile in a large study involving 34 575 cirrhosis patients, deep learning models including deep neural networks, outperformed the traditional MELD score in predicting mortality at various time frames (90, 180, and 365 days).17
In the current study, the superior performance of traditional ML models like Logistic Regression in comparison to the newer deep learning models such as multilayer ANN, RNNs, and LSTM could be attributed to several factors. First, with small sample sizes (173 in our case), traditional machine learning models are reported to be often more efficient in processing and more accurate in extracting meaningful patterns from data.24 Deep learning models require a larger amount of data to capture the complexity of the problem adequately. Additionally, traditional ML models have a simpler architecture and fewer hyperparameters to tune, making them less prone to overfitting in small datasets.25
In our study, feature importance analysis revealed the top 10 contributing features for mortality prediction in patients with chronic liver disease and cirrhosis [Figure 1]. Among these features, ICU admission was found to be the most influential, followed by mechanical ventilation, mean arterial pressure, platelet count, alkaline phosphatase, bilirubin, and white blood cell count. The AI and ML models used in a previous study identified alkaline phosphatase, alanine aminotransferase, and hemoglobin as top contributing features, besides MELD-Na variables in predicting mortality in patients with liver cirrhosis.17
Our findings and those from previous studies suggest that clinical indicators of disease severity and organ dysfunction can serve as important indicators for mortality prediction in this patient population. They provide additional factors associated with short-term poor outcomes which were not highlighted in traditional prognostic scores (i.e MELD, CLIF-C),9,26 and may further empower clinicians to make well-informed decisions regarding care prioritization, including the advisability of liver transplantation or a re-evaluation of appropriateness of certain treatments.
It is important to note the limitations of our study. First, our analysis was based on a relatively small dataset, from a single center, on a mostly ethnically homogeneous (Omani) cohort. These factors may impact the generalizability of our results. Future studies with larger and more diverse datasets in different populations are warranted to validate the performance of these models and to improve them. Additionally, external validation of the developed models using independent cohorts would provide further evidence of the models’ real-life applicability. Furthermore, our study focused on predicting mortality within a 28-day period, and longer-term predictions were not explored. Future research could use these models on different prediction horizons to enhance their clinical utility.
Conclusion
This study demonstrates the potential of ML and deep learning techniques in predicting 28-day mortality in patients admitted with acute decompensation of chronic liver disease. The Logistic Regression, Naïve Bayes, and Random Forest models showed favorable performance metrics, indicating their utility in mortality prediction. The top predictive features for 28-day mortality included ICU admission, mechanical ventilation, and mean arterial pressure. Implementing these models in clinical practice may enhance risk stratification and aid in timely intervention for patients with chronic liver disease and cirrhosis for optimal outcomes. More studies using larger cohorts, external datasets, and longer time frames are necessary to establish the robustness and generalizability of these models.
Disclosure
The authors declared no conflict of interest. No funding was received for this study.
references
- 1. Devarbhavi H, Asrani SK, Arab JP, Nartey YA, Pose E, Kamath PS. Global burden of liver disease: 2023 update. J Hepatol 2023 Aug;79(2):516-537.
- 2. Al Kaabi H, Al Alawi AM, Al Falahi Z, Al-Naamani Z, Al Busafi SA. Clinical Characteristics, Etiology, and Prognostic Scores in Patients with Acute Decompensated Liver Cirrhosis. J Clin Med 2023 Sep;12(17):5756.
- 3. Mahmud N. Selection for Liver Transplantation: Indications and Evaluation. Curr Hepatol Rep 2020;19(3):203-212.
- 4. Bajaj JS, Choudhury AK, Xie Q, Kamath PS, Topazian M, Hayes PC, et al; CLEARED Investigators. Global disparities in mortality and liver transplantation in hospitalised patients with cirrhosis: a prospective cohort study for the CLEARED Consortium. Lancet Gastroenterol Hepatol 2023 Jul;8(7):611-622.
- 5. Cucchetti A, Vivarelli M, Heaton ND, Phillips S, Piscaglia F, Bolondi L, et al. Artificial neural network is superior to MELD in predicting mortality of patients with end-stage liver disease. Gut 2007 Feb;56(2):253-258.
- 6. Trebicka J, Sundaram V, Moreau R, Jalan R, Arroyo V. Liver Transplantation for Acute-on-Chronic Liver Failure: Science or Fiction? Liver Transpl 2020 Jul;26(7):906-915.
- 7. Peng Y, Qi X, Guo X. Child-pugh versus MELD score for the assessment of prognosis in liver cirrhosis: a systematic review and meta-analysis of observational studies. Medicine (Baltimore) 2016 Feb;95(8):e2877.
- 8. Barosa R, Roque Ramos L, Patita M, Nunes G, Fonseca J. CLIF-C ACLF score is a better mortality predictor than MELD, MELD-Na and CTP in patients with acute on chronic liver failure admitted to the ward. Rev Esp Enferm Dig 2017 Jun;109(6):399-405.
- 9. Ramzan M, Iqbal A, Murtaza HG, Javed N, Rasheed G, Bano K. Comparison of CLIF-C ACLF score and MELD score in predicting ICU mortality in patients with acute-on-chronic liver failure. Cureus 2020 Feb;12(2):e7087.
- 10. Marot A, Henrion J, Knebel JF, Trépo E, Moreno C, Deltenre P. A model for individualized prediction of liver-related death in outpatients with alcohol-associated cirrhosis. Hepatol Commun 2023 Aug;7(9):e0229.
- 11. Zanetto A, Campello E, Bulato C, Gavasso S, Farinati F, Russo FP, et al. Increased platelet aggregation in patients with decompensated cirrhosis indicates higher risk of further decompensation and death. J Hepatol 2022 Sep;77(3):660-669.
- 12. Thuluvath PJ, Alukal JJ, Zhang T. A model to predict inhospital mortality in patients with cirrhosis, ascites and hyponatremia. Eur J Gastroenterol Hepatol 2022 Jun;34(6):591-597.
- 13. Angraal S, Mortazavi BJ, Gupta A, Khera R, Ahmad T, Desai NR, et al. Machine learning prediction of mortality and hospitalization in heart failure with preserved ejection fraction. JACC Heart Fail 2020 Jan;8(1):12-21.
- 14. Bhat M, Rabindranath M, Chara BS, Simonetto DA. Artificial intelligence, machine learning, and deep learning in liver transplantation. J Hepatol 2023 Jun;78(6):1216-1233.
- 15. Banerjee R, Das A, Ghoshal UC, Sinha M. Predicting mortality in patients with cirrhosis of liver with application of neural network technology. J Gastroenterol Hepatol 2003 Sep;18(9):1054-1060.
- 16. Cross SS, Harrison RF, Kennedy RL. Introduction to neural networks. Lancet 1995 Oct;346(8982):1075-1079.
- 17. Guo A, Mazumder NR, Ladner DP, Foraker RE. Predicting mortality among patients with liver cirrhosis in electronic health records with machine learning. PLoS One 2021 Aug;16(8):e0256428.
- 18. Hou Y, Zhang Q, Gao F, Mao D, Li J, Gong Z, et al. Artificial neural network-based models used for predicting 28- and 90-day mortality of patients with hepatitis B-associated acute-on-chronic liver failure. BMC Gastroenterol 2020 Mar;20(1):75.
- 19. Al-Yarabi A, Al Balushi H, Al Hatmi K, Al Yahyaie R, Al Alawi AM, Al Zeedy K, et al. Inappropriate hospital stay of patients admitted under care of general medicine units: a retrospective study. Sultan Qaboos Univ Med J 2023 May;23(2):174-181.
- 20. Al Sibani M, Al-Maqbali JS, Yusuf Z, Al Alawi AM. Incidence and risk factors for 28 days hospital readmission: a retrospective study from Oman. Oman Med J 2022 Sep;37(5):e423.
- 21. Sarker IH. Machine learning: algorithms, real-world applications and research directions. SN Comput Sci 2021;2(3):160.
- 22. Choi RY, Coyner AS, Kalpathy-Cramer J, Chiang MF, Campbell JP. Introduction to machine learning, neural networks, and deep learning. Transl Vis Sci Technol 2020 Feb;9(2):14.
- 23. Hu C, Anjur V, Saboo K, Reddy KR, O’Leary J, Tandon P, et al. Low predictability of readmissions and death using machine learning in cirrhosis. Am J Gastroenterol 2021 Feb;116(2):336-346.
- 24. Maniar KM, Lassarén P, Rana A, Yao Y, Tewarie IA, Gerstl JV, et al. Traditional machine learning methods versus deep learning for meningioma classification, grading, outcome prediction, and segmentation: a systematic review and meta-analysis. World Neurosurg 2023 Nov;179:e119-e134.
- 25. Gupta R, Srivastava D, Sahu M, Tiwari S, Ambasta RK, Kumar P. Artificial intelligence to deep learning: machine intelligence approach for drug discovery. Mol Divers 2021 Aug;25(3):1315-1360.
- 26. Shi Y, Shu Z, Sun W, Yang Q, Yu Y, Yang G, et al. Risk stratification of decompensated cirrhosis patients by chronic liver failure consortium scores: classification and regression tree analysis. Hepatol Res 2017 Mar;47(4):328-337.