Myeloid leukemia is a heterogeneous group of diseases characterized by infiltration of the blood, bone marrow, and other tissues by neoplastic cells of the hematopoietic system.1 In 2022, 20 050 new cases of acute myeloid leukemia (AML) were estimated in the USA.2 Data demonstrated an association of myeloid leukemia with irradiation, smoking, some rare congenital abnormalities, chemical exposure, and obesity.3
According to the World Health Organization classification of hematological malignancies, recurrent genetic abnormalities were identified in AML. These genetic abnormalities are associated with distinctive clinicopathological features and have prognostic significance. The Cancer Genome Atlas Research Network evaluation of 200 AML cases found an average of 13 mutations per AML case, with at least 23 recurrent mutations identified. The most common identified mutations are FLT3, NPM1, CEBPA, cKIT, NRAS, MLL, WT1, IDH1/2, TET2, DNMT3A, and ASXL1.
In recent years, the analysis of whole genome led to the identification of two mutually exclusive mutations in isocitrate dehydrogenase IDH1 genes: IDH1 in the cytoplasm and IDH2 in the mitochondria.4,5
After the discovery of IDH1 and IDH2, various studies were conducted to evaluate the prevalence of these mutations and their clinical and prognostic impact. Three main loci were studied: IDH1R132, IDH2R140, and IDH2R172.
Studies reported IDH mutations in around 33% of patients, including 6–16% for IDH1 and 8–19% for IDH2 mutations.6-8 Five missense mutations were found in IDH1, including R132H, R132C, R132S, R132G, and R132V. IDH1R132 mutation was associated with other concurrent mutations including NPM1, FLT3, CEBPA, and NRAS.9 Moreover, IDH2 mutations were found in 10.4%, with two missense mutations detected: R140Q and R172K.10
IDH mutation has a significant association with old age and higher median platelet counts, normal karyotype AML, cytogenetics intermediate-risk group, and NPM1 and FLT3-ITD mutations.6,7,11,12 IDH1 mutation status is an unfavorable prognostic factor11 and patients harboring IDH mutations have a significantly lower rate of five-year overall survival (OS) of 15.6% than patients who lack the IDH mutation (32.0%).7
The remission rates by AML treatment status were reported as 68% during induction, 42% in Salvage-1 (S1), and 27% in Salvage-2 and beyond (S21).12 In addition, no difference in response identified by IDH mutation status was found.12
All the previous studies have addressed the impact and clinicopathological features of IDH mutations in AML patients conducted internationally, and no local studies are available. In addition to the common gene mutations detected frequently in AML patients such as NPM1, FLT3, and others where analysis at hematology department at Sultan Qaboos University Hospital (SQUH), the IDH gene mutation testing will serve as an additional important diagnostic molecular marker in Omani patients with AML. To fill the gap, our study aimed to estimate the prevalence of IDH mutations among Omani patients diagnosed with AML and the common types of IDH mutation will be identified and correlated with the clinical and laboratory findings.
Methods
This single-center retrospective cohort study was conducted in SQUH. We used archived DNA collected from October 2009 to October 2019. The clinical data of the study population were obtained from the hospital information system. This research was ethically approved by the Institutional Review Board (IRB) at SQU (MREC #2048) and confidentiality was protected.
We included all Omani patients (pediatric and adult) treated at SQUH who were diagnosed with AML and had diagnostic DNA available for testing. All other patients who have missing data or their DNA extraction done at relapse or for other diagnosis (i.e., myelodysplastic syndrome) were excluded from the study.
The following hematological parameters have been assessed in all patients; full blood count including hemoglobin, hematocrit, white blood cells (WBC), and platelets count. Additionally, blood film smear, bone marrow aspirate, karyotyping, and DNA-based genetic studies were evaluated.
For IDH1 and IDH2 mutations detection, genomic DNA was extracted from EDTA-anticoagulated whole blood using the QIAamp DNA blood mini kit (Qiagen Inc, Hilden, Germany). The concentration and quality of the sample DNA were checked by NanoDrop ND-1000 (Nano-Drop Technologies, Wilmington, USA). The target regions for IDH1 and IDH2 genes were polymerase chain reaction amplified and directly sequenced using 3500 Genetic Analyzer. FLT3-ITD and NPM1 mutations were screened by capillary electrophoresis as described before.
Statistical analysis was conducted using SPSS (IBM Corp. Released 2019. IBM SPSS Statistics for Windows, Version 26.0. Armonk, NY: IBM Corp). The Kaplan-Meier method and the log-rank test were utilized to estimate the distribution of OS, with a p-value of < 0.05 was considered statistically significant. Continuous variables were presented as median (IQRs), while categorical variables such as French American-British classification and cytogenetics were presented as frequency.
Results
We analyzed a total of 61 patients with AML, with a median age of 40 years. The baseline characteristics of the study population are summarized in Table 1.

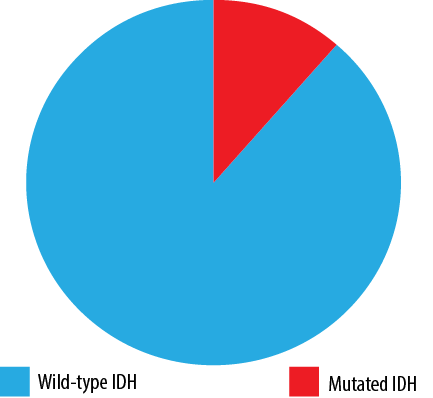 Figure 1: Percentage of IDH mutations.
Figure 1: Percentage of IDH mutations.

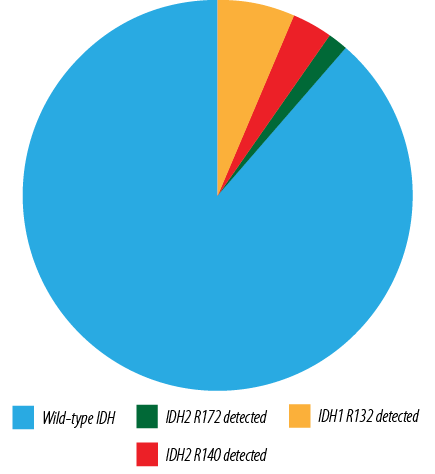 Figure 2: Loci of IDH1/IDH2 mutations detected.
Figure 2: Loci of IDH1/IDH2 mutations detected.
Table 1: Patient demographic features (N = 61).
|
Age at diagnosis, median (IQR), years
|
40 (25.5–65.5)
|
|
Gender
|
|
|
Male
|
31 (50.8)
|
|
Female
|
30 (49.2)
|
|
Hemoglobin, median (IQR), g/dL
|
8.3 (7.4–9.8)
|
|
WBC, median (IQR), 109/L
|
9.8 (2.9–53.8)
|
|
PLT count, median (IQR), 109/L
|
35 (23–99)
|
|
Blast %, median (IQR)
|
77 (52.3–87.2)
|
|
LDH, median (IQR)
|
394 (266–857)
|
|
Flow cytometry results
|
|
|
APML
|
10 (16.4)
|
|
AML
|
46 (75.4)
|
|
Mixed phenotype
|
5 (8.2)
|
|
Risk stratification
|
|
|
Favorable prognosis
|
8 (13.1)
|
|
Intermediate risk
|
17 (27.9)
|
|
High risk
|
35 (57.4)
|
|
Study group status
|
|
|
Alive
|
17 (27.9)
|
|
Died
|
38 (62.3)
|
WBC: white blood cell; PLT: platelet; LDH: lactate dehydrogenase;
APML: acute promyelocytic leukaemia; AML: acute myeloid leukemia.
Out of all tested AML cases, 54 (88.5%) had no IDH mutations (IDH1/IDH2 wild-type AML), while seven (11.5%) harbored either an IDH1 or an IDH2 mutation [Figure 1]. Among all tested AML patients, an IDH1 mutation was detected in four (6.6%) cases, and an IDH2 mutation was detected in three cases (4.9%). The main loci identified in IDH1 mutation were p.R132C in three cases (75.0%) and p.R132H in one case (25.0%). For IDH2 mutation, p.R140Q and p.R172K were detected in 3.3% and 1.6%, respectively [Figure 2]. No cases had mutations in both IDH1 and IDH2, suggesting that these mutations are mutually exclusive.
The median age of the wild-type group was 37 years, whereas the median age for the mutated IDH group was 56 years. The female-to-male ratio in the IDH mutated group was 2.5:1. The mutated IDH group tended to have a lower WBC with a median of 4.4 × 109/L (1–37 × 109/L) and a higher platelet count with a median of 47 × 109/L (32–138 × 109/L) at diagnosis. There was no difference in hemoglobin levels between the two groups, with a median of 8.3 g/dL for the wild-type IDH and 7.9 g/dL for the mutated IDH group. Patient characteristics of both the wild-type and mutated IDH are displayed in Table 2.
Table 2: Comparison of patient demographic features between the study subgroups.
|
Age at diagnosis, median (IQR), years
|
|
Gender, n (%)
|
|
|
|
Male
|
|
Female
|
|
Hemoglobin, median (IQR), g/dL
|
|
WBC, median (IQR), 109/L
|
|
PLT count, median (IQR), 109/L
|
|
Blast %, median (IQR)
|
|
LDH, median (IQR)
|
|
Flow cytometry results, n (%)
|
|
|
|
APML
|
|
AML
|
|
Mixed phenotype
|
|
Risk stratification, n (%)
|
|
|
|
Favorable prognosis
|
|
Intermediate risk
|
|
High risk
|
|
Study group status, n (%)
|
|
|
|
Alive
|
|
Died
|
WBC: white blood cell; PLT: platelet; LDH: lactate dehydrogenase; APML: acute promyelocytic leukaemia; AML: acute myeloid leukemia.
The median blast count in the mutated IDH group was 81% (63–90%) whereas in the wild-type IDH group, it was 77.0% (51.5–85.7%). All the cases in the mutated group were classified as AML not otherwise specified based on World Health Organization AML classification. These cases were further classified using French American-British classification into AML without maturation (M1) in one patient (14.3%), AML with maturation (M2) in one patient (14.3%), acute promyelocytic leukemia variant (M3-v) in one patient (14.3%), acute myelomonocytic leukemia (M4) in two patients (28.6%), and acute monocytic leukemia (M5) in two patients (28.6%). Dysplastic features were described in four patients (57.1%) with mutated IDH. These dysplastic features were identified mainly in granulocytes in one patient (14.3%), megakaryocytes in two patients (28.6%), and erythroid lineages in one patient (14.3%) [Table 3].
Table 3: Cytogenetic and molecular features of AML with IDH1R132, IDH2R140, and IDH2R172 mutations.
|
74/F
|
M4
|
N. karyotype
|
Intermediate
|
IDH1
|
CGT-TGT
|
p.R132C
|
NPM1-, FLT3-ITD-
|
|
73/F
|
M1
|
+4
|
Intermediate
|
IDH1
|
CGT-TGT
|
p.R132C
|
NPM1+, FLT3-ITD-
|
|
56/M
|
M5
|
+11
|
Intermediate
|
IDH1
|
CGT-TGT
|
p.R132C
|
NPM1-, FLT3-ITD-
|
|
36/M
|
M5
|
-Y
|
Intermediate
|
IDH1
|
CGT-TGT
|
p.R132H
|
NPM1-, FLT3-ITD-
|
|
70/F
|
M2
|
T(10;17)
|
Intermediate
|
IDH2
|
CCG-CAG
|
p.R140Q
|
NPM1-, FLT3-ITD-
|
|
48/F
|
M2
|
N. karyotype
|
Intermediate
|
IDH2
|
CCG-CAG
|
p.R140Q
|
NPM1-, FLT3-ITD-
|
AML: acute myeloid leukemia; IDH, isocitrate dehydrogenase; F: female; M: male; N: normal. FAB: French-American-British classification of AML.
Among the studied AML patients, favorable, intermediate, and adverse cytogenetic risk was found in eight (13.1%), 17 (27.9%), and 35 (57.4%) cases, respectively. In the mutated group, two (28.6%) AML cases had normal karyotype, while five cases (71.4%) exhibited various cytogenetic abnormalities including trisomy 4, trisomy 11, trisomy 8, del (Y), and t(10,13) All IDH mutated cases (100%) were in the intermediate-risk cytogenetics group. Furthermore, 14.3% (one patient) in the IDH mutated group displayed concurrent mutations of both IDH1 and NPM1 [Table 4]. Unfortunately, the assessment of associations with other molecular abnormalities was hindered by the small sample size.
Table 4: Hematologic and morphologic features of AML with IDH1R132, IDH2R140, and IDH2R172 mutations.
|
1
|
IDH1
|
74/F
|
M4
|
3
|
10.1
|
138
|
31
|
++
|
Myeloid, Megas
|
|
2
|
IDH1
|
73/F
|
M1
|
91
|
8.3
|
338
|
81
|
-
|
Megas
|
|
3
|
IDH1
|
56/M
|
M5
|
1
|
7.2
|
32
|
79
|
?
|
?
|
|
4
|
IDH1
|
36/M
|
M5
|
8
|
5.9
|
14
|
63
|
-
|
Absent
|
|
5
|
IDH2
|
70/F
|
M2
|
4
|
7.9
|
47
|
90
|
-
|
Absent
|
|
6
|
IDH2
|
48/F
|
M2
|
37
|
6.9
|
35
|
95
|
++
|
Absent
|
AML: acute myeloid leukemia; IDH, isocitrate dehydrogenase. FAB: French-American-British classification of AML. WBC: white blood cell; Hb: hemoglobin; PLT: platelet; F: female; M: male. Case 3: Bone marrow aspiration not done as the patient refused.
The median OS for the IDH1 and IDH2 mutated groups was nine months (p = 0.593). The estimated OS in the study cohort did not significantly differ between IDH wild-type and mutated IDH [Figure 3].

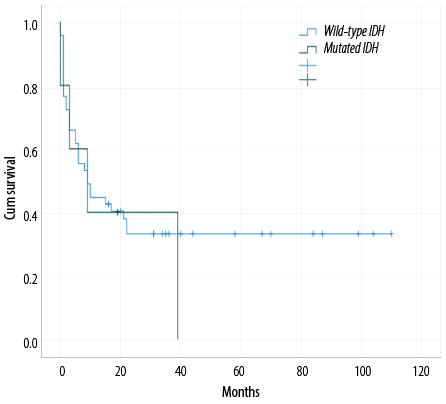 Figure 3: Overall survival between wild-type IDH and mutated IDH.
Figure 3: Overall survival between wild-type IDH and mutated IDH.
Discussion
IDH mutations have been identified in a variety of malignancies, including gliomas,14 chondrosarcomas,15 cholangiocarcinomas,6 and breast carcinomas.7 Furthermore, these mutations have been found in myeloid neoplasms such as AML, myelodysplastic syndrome, and myeloproliferative neoplasms.3 The normal function of IDH enzymes, encoded by IDH genes, is to convert isocitrate to α-ketoglutarate. Whereas mutant IDH enzymes, reduce α-ketoglutarate to its oncometabolite 2-hydroxyglutarate. The later inhibits α-ketoglutarate-dependent enzymes such as TET2. This results in abnormal epigenetic modifications that hinder cell differentiation.8
In our retrospective study, the overall frequency of IDH mutations among AML patients was 11.5%, which is lower than the reported incidence ranging from 16% to 33%.4,5,13,16,17 This difference could be attributed to the extremely small sample size compared to other published studies. In our subgroup analysis of IDH mutations, IDH1 mutation was detected in four patients (6.6%) with the three main loci IDH1R132C (75.0%) and IDH1R132H (25.0%). Whereas IDH2 mutation was found in 4.9% of cases, with the main loci IDH2R140Q (3.3%) and IDH2R172K (1.6%). The reported incidence of IDH1 mutation ranges from 6–14% of cases while IDH2 is 8–19%.4,5,13,16,17 Hence, the incidence rates of IDH1 and IDH2 mutation were consistent with international studies.
Our study revealed that patients with IDH mutations tended to be older, with a female predominance. The median age of mutated IDH patients reported in other studies ranged from 49 to 67 years.11,16-18 Patel et al,18 reported a male-to-female ratio of 1:3 whereas Green et al,11 reported a ratio of 1:2. A lower median white cell count with higher median platelet count and blast percentage were observed in IDH mutated patients at diagnosis, which is consistent with other published international studies.5,16,17 In agreement with other studies,5,18,19 all mutated IDH cases were associated with intermediate cytogenetics risk. Chotirat et al,17 identified 55% (11 cases) and 50% (12 cases) normal karyotype in mutated IDH1 and IDH2, respectively. In addition, they reported various cytogenetics abnormalities in aberrant karyotype including del(9q), trisomy 8, trisomy 11, del(12) (p12.1p13.1), t(15,17), and t(8;21). However, in this study, only 28.6% (two patients) has normal karyotype, one in each group. The identified cytogenetics abnormalities include trisomy 4, trisomy 11, trisomy 8, del (Y) and t(10,17). Furthermore, multiple studies were conducted to assess the concurrent presence of other gene mutations along with IDH mutation. Patel et al,18 reported the following additional gene mutations including NPM1, FLT3-ITD, CEBPA, NRAS, KIT, and FLT3-D835 in IDH1-mutated cases. A significant association between NPM1 and mutated IDH1 (74%; p < 0.001) and IDH2 (60%; p < 0.001) was demonstrated by Green et al.11 We found concurrent presence of NPM1 mutation in one case with mutated IDH1. Unfortunately, our study’s small sample size prevented a comprehensive assessment of such associations. Table 5 compares our results with other international studies.
Table 5: Comparison of results between our study and other international results for patients with isocitrate dehydrogenase 1 (IDH1).
|
IDH1
|
4/61
|
16/188
|
27/493
|
49/358
|
30/275
|
|
Age, years
|
61.5
|
48.9
|
52.5
|
62
|
50
|
|
(Mean, range)
|
(43.5–70.1)
|
|
(25–75)
|
(21–82)
|
(33–80)
|
|
Female:male
|
3:1
|
1.3:1
|
1.1:1
|
1:1.1
|
1:1.7
|
|
% Blast (mean, range)
|
80 (55–91.5)
|
76.7 ± 16.4
|
NA
|
73 (33–99)
|
80 (20–99)
|
|
Risk stratification, n (%)
|
|
|
|
NA
|
NA
|
|
Favorable
|
0 (0.0)
|
0
|
0
|
|
|
|
Intermediate
|
4 (100)
|
16 (100)
|
26/26 (100)
|
|
|
|
High risk
|
0 (0.0)
|
0
|
0
|
|
|
|
IDH1 mutation, n (%)
|
|
|
|
|
|
|
R132H
|
1 (25.0)
|
7 (44.0)
|
7 (26.0)
|
24 (49.0)
|
21 (70.0)
|
|
R132C
|
3 (75.0)
|
8 (50.0)
|
10 (37.0)
|
15 (31.0)
|
5 (17.0)
|
|
R132S
|
-
|
1 (6.0)
|
5 (19.0)
|
5 (10.0)
|
3 (10.0)
|
|
R132L
|
-
|
-
|
1 (4.0)
|
-
|
-
|
|
R132G
|
-
|
-
|
4 (15.0)
|
-
|
1 (3.0)
|
|
Coexisting mutations, n (%)
|
|
|
|
|
|
NPM1
|
1 (25.0)
|
7 (44.0)
|
15 (56.0)
|
34 (71.0)
|
17 (57.0)
|
|
FLT3-ITD
|
-
|
4 (25.0)
|
10 (37.0)
|
10 (20.0)
|
4 (13.0)
|
** Meta-analysis data extracted from acute myeloid leukemia with IDH1 or IDH2 Mutations: Frequency and Clinicopathologic Features study, Am J Clin Pathol. 2011 Jan; 135 (1): 35-45.
In the context of OS, our study did not find a statistically significant impact of IDH mutations on survival compared to wild-type IDH. However, the small sample size may have limited our ability to detect a significant difference. This study represents the first effort to investigate the prognostic significance of IDH mutations in AML patients in Oman. Nonetheless, the study has several limitations, including its observational, retrospective nature, single-center design, and a limited number of AML patients with IDH mutations. The effectiveness of the current therapy to clear such mutation could not be assessed due to the short survival of patients. Future studies with larger sample sizes from multiple centers (including Royal Hospital and Oman’s National Genetics Center) may provide more robust insights into the prognostic implications of IDH mutations and their response to therapy.
Conclusion
Our study revealed that the overall frequency of IDH mutations among AML patients in our cohort was 11.5%, with IDH1 mutations accounting for 6.6% and IDH2 mutations for 4.9%. Patients with IDH mutations tended to be older in age, exhibit a lower median WBC, higher median platelet count, and a higher blast percentage at diagnosis. Additionally, mutated IDH was associated with the intermediate cytogenetic risk group. However, we did not find a significant prognostic impact of IDH mutations on survival between the study groups. Considering these findings and the availability of new, targeted therapies for IDH mutations, we recommend implementing testing for IDH mutations as a primary diagnostic test for all newly diagnosed AML cases. This approach could facilitate more personalized treatment strategies, taking into account the specific molecular characteristics of each patient’s disease.
Disclosure
The authors declared no conflicts of interest. No funding was received for this study.
Acknowledgments
We would like to thank Mr. Mohammed Al Rawahi, Senior Biomedical Scientist, Molecular Biology Section, SQUH for his support in molecular testing and analysis of IDH mutation.
references
- 1. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al, eds. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Revised 4th edition. International Agency for Research on Cancer (IARC) 69372 Lyon Cedex 08, France; 2017.
- 2. Cancer Facts & Figures 2022. Acute myeloid leukemia [cited 2023 March 3]. Cancer.org. 2022. Available from: https://www.cancer.org/content/dam/CRC/PDF/Public/8677.00.pdf
- Platt MY, Fathi AT, Borger DR, Brunner AM, Hasserjian RP, Balaj L, et al. Detection of Dual IDH1 and IDH2 Mutations by Targeted Next-Generation Sequencing in Acute Myeloid Leukemia and Myelodysplastic Syndromes. J Mol Diagn 2015 Nov;17(6):661-668.
- Abbas S, Lugthart S, Kavelaars FG, Schelen A, Koenders J, Zeilemaker A, et al. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia (AML). Blood 2010 Sep;116(12):2122-6.
- DiNardo CD, Ravandi F, Agresta S, Konopleva M, Takahashi K, Kadia T, et al. Characteristics, clinical outcome, and prognostic significance of IDH mutations in AML. Am J Hematol 2015 Aug;90(8):732-736.
- Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist 2012;17(1):72-79.
- Fathi AT, Sadrzadeh H, Comander AH, Higgins MJ, Bardia A, Perry A, et al. Isocitrate dehydrogenase 1 (IDH1) mutation in breast adenocarcinoma is associated with elevated levels of serum and urine 2-hydroxyglutarate. Oncologist 2014 Jun;19(6):602-607.
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010 Dec;18(6):553-567.
- Yamaguchi S, Iwanaga E, Tokunaga K, Nanri T, Shimomura T, Suzushima H, et al. IDH1 and IDH2 mutations confer an adverse effect in patients with acute myeloid leukemia lacking the NPM1 mutation. Eur J Haematol 2014 Jun;92(6):471-477.
- Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Krönke J, Bullinger L, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol 2010 Aug;28(22):3636-3643.
- Green CL, Evans CM, Hills RK, Burnett AK, Linch DC, Gale RE. The prognostic significance of IDH1 mutations in younger adult patients with acute myeloid leukemia is dependent on FLT3/ITD status. Blood 2010 Oct;116(15):2779-2782.
- Aref S, Kamel Areida S, Abdel Aaal MF, Adam OM, El-Ghonemy MS, El-Baiomy MA, et al. Prevalence and Clinical Effect of IDH1 and IDH2 Mutations Among Cytogenetically Normal Acute Myeloid Leukemia Patients. Clin Lymphoma Myeloma Leuk 2015 Sep;15(9):550-555.
- Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Krönke J, Bullinger L, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol 2010 Aug;28(22):3636-3643.
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009 Feb;360(8):765-773.
- Kerr DA, Lopez HU, Deshpande V, Hornicek FJ, Duan Z, Zhang Y, et al. Molecular distinction of chondrosarcoma from chondroblastic osteosarcoma through IDH1/2 mutations. Am J Surg Pathol 2013 Jun;37(6):787-795.
- Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrózek K, Margeson D, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. J Clin Oncol 2010 May;28(14):2348-2355.
- Chotirat S, Thongnoppakhun W, Promsuwicha O, Boonthimat C, Auewarakul CU. Molecular alterations of isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) metabolic genes and additional genetic mutations in newly diagnosed acute myeloid leukemia patients. J Hematol Oncol 2012 Mar;5(1):5.
- Patel KP, Ravandi F, Ma D, Paladugu A, Barkoh BA, Medeiros LJ, et al. Acute myeloid leukemia with IDH1 or IDH2 mutation: frequency and clinicopathologic features. Am J Clin Pathol 2011 Jan;135(1):35-45.
- Green CL, Evans CM, Zhao L, Hills RK, Burnett AK, Linch DC, et al. The prognostic significance of IDH2 mutations in AML depends on the location of the mutation. Blood 2011 Jul;118(2):409-412.