In most parts of the world, especially developing countries including Nigeria, the detection of resistant genes and/or multidrug-resistant (MDR) enzymes responsible for the negative response of pathogenic bacteria to potent antimicrobial onslaught is still ill-detected in our hospitals. This is because routine antimicrobial susceptibility studies (as is obtainable in most of our hospitals) is ineffective in detecting these MDR organisms. Gram-negative bacteria represent the most relevant reservoir of resistance to antibiotics in the environment. The natural selection of resistant clones of bacteria in the environment by antimicrobial selective pressure is a relevant mechanism for spreading antibiotic resistance traits in the community and hospital environment.1,2 This is in scenarios where antimicrobials are used irrationally, and even in the propagation of livestock, poultry birds, and other veterinary purposes. AmpC β-lactamases are cephalosporinases that belong to molecular class C β-lactamases and are encoded on the chromosomes of many gram-negative bacteria.1–3 The genes encoding AmpC β-lactamases are much more frequently chromosomal than plasmid-mediated. The AmpC β-lactamases are clinically important β-lactamases because they confer antimicrobial resistance to the narrow-spectrum, expanded-spectrum, and the broad-spectrum cephalosporins including cefotaxime, ceftazidime, ceftriaxone, aztreonam, and the penicillins. Resistance is also expressed towards β-lactamase inhibitors such as amoxicillin-clavulanic acid.3,4 Most of the genera of the family Enterobacteriaceae produce AmpC enzymes through an inducible mechanism in which case the presence of broad-spectrum antibiotics sparks enzyme production in the organism.4,5 The production of some MDR enzymes such as extended-spectrum beta-lactamases (ESBLs) by gram-negative bacteria can be masked by the coexpression of AmpC in the same organism.6,7 One of the biggest challenges facing the health sector across the globe, especially in the area of infection control and prevention, is in the adequate containment of multidrug-resistant gram-negative organisms (MDRGNOs). It is worrisome that MDRGNOs are not restricted to the hospital environment and bacterial pathogens including Klebsiella species, Pseudomonas aeruginosa, and Escherichia coli, which produce enzymes that degrade some of the world’s potent and effective drugs such as carbapenems and cephalosporins, are now found in the community.8–12 Measuring the prevalence of these organisms in both hospital and community settings coupled with proper use of available drugs and susceptibility testing of these organisms before the prescription or use of some expanded-spectrum drugs (especially in hospital environments) is critical to reducing the possible risks associated with infections due to MDRGNOs.6–12
Food-producing animals harboring MDR genes together with genes that mediate the production of some high-profile antibiotic hydrolyzing enzymes such as AmpC enzymes possess health risks to the human population due to their potential contribution in the spread of drug-resistant microorganisms in the community. We used multiplex polymerase chain reaction (PCR) to detect the occurrence of FOX-1 genes in E. coli isolates from abattoir sources in Abakaliki, Nigeria.
Methods
In line with the World Medical Association Declaration of Helsinki on the principles for medical research involving human subjects and identifiable human and animal material/data, this research was undertaken based on a thorough knowledge of the scientific literature and adequate laboratory protocol. Oral consent was sought from the abattoir operators for the collection and processing of the anal swab samples. There was no written ethical approval as the samples were collected at the point of slaughter, and the animals were handled with respect and total care. There was minimal pain incurred by the animals during sample collection.
A total of 120 samples were collected from the anal region of cows in a local abattoir in Abakaliki, Nigeria, using sterile swab sticks. To isolate E. coli, a loopful of the turbid solution from overnight nutrient broth culture was plated onto eosin methylene blue (EMB) agar and MacConkey agar (Oxoid, UK) plates. Each plate was incubated at 30 oC for 18 hours and showed a positive reaction. Suspect colonies of E. coli was aseptically subcultured onto freshly prepared EMB and MacConkey agar plates for the isolation of pure cultures, and these were incubated at 30 oC for 18 hours. E. coli was identified using standard microbiology identification technique.8
Antimicrobial susceptibility studies were carried out using the modified Kirby-Bauer disk diffusion method on Mueller-Hinton (MH) agar plates (Oxoid, UK) as previously described.9,10 Briefly, the test antibiotic disks were aseptically placed at a distance of 15 mm apart on MH agar plates already swabbed or inoculated with the E. coli isolates (adjusted to 0.5 McFarland turbidity standards). Each plate was incubated at 30 oC for 18 hours. E. coli ATCC 25922 strain was used as the quality control strain for the antimicrobial susceptibility test. Inhibition zones were interpreted according to Clinical and Laboratory Standard Institute (CLSI) criteria.
To characterize β-lactamase the impregnated end of the nitrocefin stick was used to pick the test isolate; and the stick was macroscopically observed after 5–15 minutes for a color change.11 A positive reaction was shown by the development of a pink-red color while a negative reaction was indicated by no color change.
Bacterial strains that produce AmpC β-lactamase are resistant to the cephamycins but susceptible to the fourth generation cephalosporin, cefepime.12,13 All test isolates were subjected to the antimicrobial activity of cefoxitin disk (30 µg) on aseptically streaked MH agar plates. Each plate was incubated at 30 oC for 18 hours.10,12 Inhibition zones were interpreted according to CLSI criteria.
Cefoxitin-cloxacillin double-disk synergy test (CC-DDST) was performed according to the method described by Polsfuss et al.14 Single disks containing 30 µg of cefoxitin were placed 20 mm away from a disk containing 200 µg of cloxacillin on MH agar plates already inoculated with the test bacteria (equivalent to 0.5 McFarland turbidity standards). Each plate was incubated at 30 oC for 18 hours. Inhibition zones were interpreted according to CLSI criteria. A difference of 4 mm in the cefoxitin-cloxacillin inhibition zones minus the cefoxitin disk used alone was indicative of AmpC enzyme production phenotypically.
The presence of AmpC β-lactamase genes in the E. coli isolates was investigated using a multiplex PCR assay targeting the FOX-1 AmpC β-lactamase genes as described by Perez-Perez and Hanson.15 Each of the E. coli isolates was inoculated from nutrient agar plates (Oxoid, UK) into 5 mL of peptone water (Oxoid, UK). The inoculated broths were incubated at 30 oC for 18–24 hours with mild shaking. Bacterial cells from the overnight broth culture were aseptically harvested by centrifugation at 5000 rpm for 5 minutes, and the supernatant was decanted. The pellets collected at the bottom of the tube were resuspended in 100 µL of Tris-EDTA (TE) buffer. The cells in the TE buffer tubes were lysed by heating at 100 oC for 10 minutes in a water bath. Finally, the solution was centrifuged at 5000 rpm for 5 minutes (to remove cellular debris). The supernatant was used as the DNA template source for gene amplification. Multiplex PCR amplification of AmpC β-lactamase genes from the test isolates was carried out using primers synthesized and supplied by Inqaba Biotechnical Industries Ltd (Inqaba Biotechnical Industries Ltd, South Africa) in a thermal cycler (Lumex Instruments, Canada) and with a final PCR mixture of 26.5 µL containing 0.2 µL of Taq polymerase enzyme U/µL, 2.5 µL of 10X PCR buffer along with 2.5 µL MgCl2, 1 µL of 10 pM from each of the forward and reverse primers, 2.5 µL of dNTPs MIX (2 Mm), 3 µL of DNA template (from the test isolates), and 14.8 µL of nuclease-free water. To determine the E. coli isolates possessing the FOX-1 AmpC β-lactamase genes, primers for FOX-1 genes were used: FOXM-Forward (AAC ATG GGG TAT CAG GGA GAT G) and FOXM-Reverse (CAA AGC GCG TAA CCG GAT TGG).15 Amplification included initial denaturation at 94 oC for 3 minutes, followed by 25 cycles of DNA denaturation at 94 oC for 30 seconds, annealing at 64 oC for 30 seconds, and extension at 72 oC for 1 minute. Gel electrophoresis of the multiplex PCR products was carried out using 1.5% agarose gel (Inqaba Biotechnical Industries Ltd, South Africa) for 2 hours at 80 V.
Results
A total of 120 swab samples were aseptically collected from the anal region of cows in a slaughterhouse in Abakaliki, Nigeria. E. coli was isolated from 48 swab samples giving a rate of 40.0% [Table 1].
Table 1: Frequency of Escherichia coli from anal swab samples.
β-lactamase enzyme production was detected in 35 (72.9%) isolates of E. coli [Table 1].
We observed a high rate of resistance among the test E. coli isolates. The E. coli isolates were highly resistant to the carbapenems, penicillins, aminoglycosides, fluoroquinolones, and cephalosporins [Figure 1]. Overall, the percentage resistance of the E. coli isolates to the tested antibiotics shows that the organism was resistant to cefoxitin (91.7%), ceftriaxone (83.3%), imipenem (85.4%), ceftazidime (87.5%), ofloxacin (81.3%), and gentamicin (85.4%). However, cefotaxime, meropenem, ampicillin, aztreonam, and cloxacillin also showed the least susceptibility to the tested isolates at the rate of 93.7%, 85.4%, 79.2%, 77.1%, and 87.5%, respectively [Figure 2].
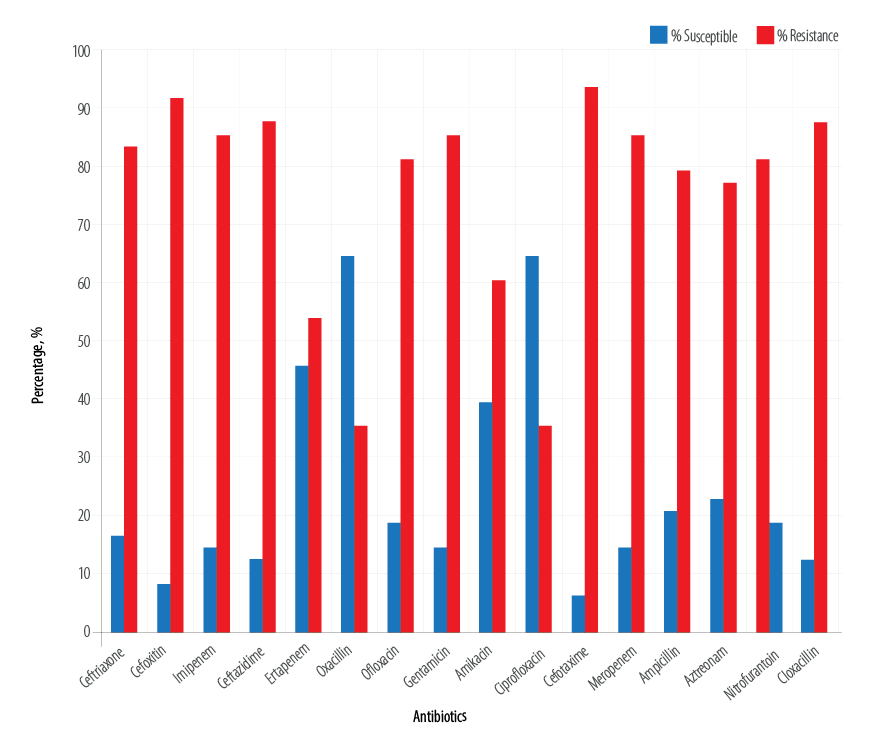
Figure 1: Susceptibility profile of the isolated Escherichia coli.
1: Ertapenem (10 µg); 2: Cefoxitin (30 µg); 3: Cefotaxime (30 µg);
4: Ciprofloxacin (10 µg); 5: Imipenem (10 µg); 6: Nitrofurantoin (10 µg);
7: Meropenem (10 µg); 8: Ampicillin (10 µg); 9: Aztreonam (30 µg);
10: Ceftriaxone (30 µg).
Figure 2: Susceptibility test plate of an Escherichia coli isolate cultured on Mueller-Hinton agar plate.
Table 2: Frequency of AmpC-producing Escherichia coli isolates.
*Cefoxitin-cloxacillin double-disk synergy test (CC-DDST).
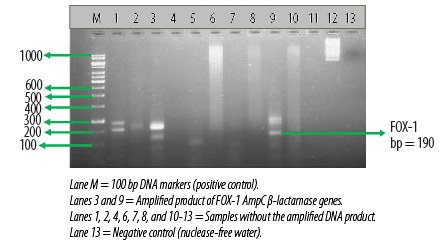
Figure 3: Electrophoretogram showing the
FOX-1 AmpC β-lactamase genes amplified from two different Escherichia coli isolates.
AmpC β-lactamase production was also detected phenotypically in six E. coli (12.5%) isolates using the CC-DDST. Table 2 also shows the result of the molecular detection of plasmid-mediated FOX-1 AmpC β-lactamase genes in the E. coli isolates by multiplex PCR. Among the 48 E. coli isolates used for this study, only three harbored the FOX-1 plasmid-mediated AmpC gene, which provokes the production of AmpC β-lactamase in the test organism. A total of 45 E. coli isolates showed no AmpC gene [Table 2]. Electrophoretic analysis of the PCR product of the test isolates after gene amplification revealed bands in the electrophoretogram, and this was positive for FOX-1 gene with a base pair size of 190 bp [Figure 3].
Discussion
The use of antibiotics in animal husbandry and other agricultural practices has contributed significantly to the burden and threat of antimicrobial resistance in the community. The improper handling and usage of animal wastes and effluents from abattoirs (inundated with antibiotic-resistant bacteria) in this part of the world necessitated our study, which investigated phenotypically and genotypically the prevalence, antibiogram, and genetic determinants of AmpC-producing E. coli from a local abattoir in Abakaliki metropolis, Ebonyi State, Nigeria. The occurrence of gram-negative bacteria including E. coli in environmental samples has been previously reported within Nigeria and in other parts of the world.11,12,16,17 Another study from Nigeria also reported that Enterobacteriaceae were the most prevalent organisms isolated from environmental samples.11 Several studies have also shown E. coli was the most prevalent gram-negative bacteria isolated from environmental samples including samples from poultry birds.16–20 The presence of β-lactamase enzyme in bacteria provides opportunity for the horizontal transmission of these enzymes from one organism to another.1,16,17,21–23 The high levels of resistance of E. coli isolates from environmental samples to some conventional antibiotics (as obtainable in this study) have been reported in the Netherlands, Nigeria, and Uganda.24–27 Bergenholtz et al,17 also reported high resistance of E. coli isolates from environmental samples to some conventional antibiotics as reported in this study. In Uganda, it was reported that 168 E. coli isolates out of 182 were resistant to several antibiotic classes including penicillins, fluoroquinolones, and aminoglycosides.25
Our study revealed that AmpC enzyme production was high in anal swab samples. In India, the prevalence of AmpC-producing E. coli isolates was 12.5%, a result similar to ours.28 However, the prevalence of AmpC enzyme-producing E. coli isolates in Iran was reported as 37.2%,29, and this result is higher than the occurrence rate of AmpC enzyme-producing E. coli isolates in this study. In the Czech Republic, two E. coli isolates from cloacal swabs of poultry birds were reported as AmpC-positive.30 The FOX gene homologs are usually the most prevalent AmpC genes aside from the CMY gene homologs found in gram-negative bacteria, which mediate their resistance to the cephamycins.3,31 The prevalence of E. coli phenotypes harboring the FOX-1 gene in this study is not in agreement with a study from Egypt, which detected the FOX-1 gene in 33.3% E. coli isolates.32 We detected the FOX-1 genes in only 6.3% of E. coli isolates. Another study from Egypt reported AmpC-producing strains in 88.46% isolates among cefoxitin resistant E. coli and Klebsiella species.33 The low percentage of FOX family-specific AmpC genes detected by the authors is in accordance with our study, which we detected by multiplex PCR.33 In Iran, the prevalence of AmpC-producing strains of E. coli was 37.2%, a result higher than ours.29 In another related study from Pakistan, it was also reported that the FOX genes were the most prevalent AmpC genes detected by PCR in E. coli isolates.34
The prevalence, emergence and spread of antibiotic-resistant bacteria in both the community and hospital environment as previously reported is notable and can impact negatively on antimicrobial therapy.33–36 In a related and recent study, it was reported that the antimicrobial armamentarium might soon be depleted due to the nefarious activities of antibiotic-resistant bacteria which have put the therapeutic effect of some drugs at risk.35
Conclusion
We report the first multiplex detection of FOX-1 AmpC genes in gram-negative bacteria in Abakaliki, Nigeria. Our results show that E. coli isolates from abattoir origin harbor antibiotic resistance genes (specifically AmpC genes) that mediate gram-negative resistance to the cephamycins. Minimizing the use of broad-spectrum antibiotics in both clinical and non-clinical settings will help to contain the emergence and spread of these organisms. The use of antibiotics to treat animals and as growth promoting agents should be curtailed and possibly replaced by proper hygienic practices and the use of vaccination to limit the emergence and spread of antibiotic-resistant bacteria in the community.
Disclosure
The authors declared no conflicts of interest. No funding was received for this study.
Acknowledgements
We appreciate the members and staff of the Biotechnology Research and Development Center of Ebonyi State University, Abakaliki for their technical guidance in carrying out the multiplex PCR technique.
references
- 1. Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrob Agents Chemother 2010 Mar;54(3):969-976.
- 2. Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother 1995 Jun;39(6):1211-1233.
- 3. Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-β-lactamases: the quiet before the storm? Clin Microbiol Rev 2005 Apr;18(2):306-325.
- 4. Shevade SU, Agrawal GN. A study on the AmpC production amongst the urinary Enterobacteriaceae isolates. J Clin Diagn Res 2013 Aug;7(8):1831-1832.
- 5. Vanwynsberghe T, Verhamme K, Raymaekers M, Cartuyvels R, Vaerenbergh KV, Boel A, et al. A Large hospital outbreak of Klebsiella pneumoniae (DHA-1 and SHV-11 Positive): Importance of detection and treatment of AmpC β-Lactamases. The Open Infectious Diseases Journal 2009;3:55-60.
- 6. Jacoby GA, Munoz-Price LS. The new β-lactamases. N Engl J Med 2005 Jan;352(4):380-391.
- 7. Fernández M, Conde S, de la Torre J, Molina-Santiago C, Ramos JL, Duque E. Mechanisms of resistance to chloramphenicol in Pseudomonas putida KT2440. Antimicrob Agents Chemother 2012 Feb;56(2):1001-1009.
- 8. Cheesbrough M. District Laboratory Practice in Tropical Countries. 2nd ed. Cambridge University Press, UK; 2006. p. 178-187.
- 9. Javeed I, Hafeez R, Anwar MS. Antibiotic susceptibility pattern of bacterial isolates from patients admitted to a tertiary care hospital in Lahore. Biomedica 2011;27:19-23.
- 10. Clinical and Laboratory Standard Institute. Performance standards for antimicrobial disk susceptibility testing. Fifteenth informational supplement, CLSI document M100-S15. USA: Wayne, PA; 2011.
- 11. Akinduti PA, Ejilude O, Motayo BO, Adeyokinu AF. Emerging multidrug resistant AmpC beta-lactamase and carbapenemase enteric isolates in Abeokuta, Nigeria. Nat Sci 2012;10(7):70-74.
- 12. Ejikeugwu C, Esimone C, Iroha I, Ugwu C, Ezeador C, Duru C, et al. Phenotypic detection of AmpC beta-lactamase among anal Pseudomonas aeruginosa isolates in a Nigerian abattoir. Archives of Clinical Microbiology 2016;7(2):1-5.
- 13. Manchanda V, Singh NP, Shamweel A, Eideh HK, Thukral SS. Molecular epidemiology of clinical isolates of ampc producing Klebsiella pneumoniae. Indian J Med Microbiol 2006 Jul;24(3):177-181.
- 14. Polsfuss S, Bloemberg GV, Giger J, Meyer V, Böttger EC, Hombach M. Practical approach for reliable detection of AmpC beta-lactamase-producing Enterobacteriaceae. J Clin Microbiol 2011 Aug;49(8):2798-2803.
- 15. Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol 2002 Jun;40(6):2153-2162.
- 16. Ben Slama K, Jouini A, Ben Sallem R, Somalo S, Sáenz Y, Estepa V, et al. Prevalence of broad-spectrum cephalosporin-resistant Escherichia coli isolates in food samples in Tunisia, and characterization of integrons and antimicrobial resistance mechanisms implicated. Int J Food Microbiol 2010 Feb;137(2-3):281-286.
- 17. Bergenholtz RD, Jørgensen MS, Hansen LH, Jensen LB, Hasman H. Characterization of genetic determinants of extended-spectrum cephalosporinases (ESCs) in Escherichia coli isolates from Danish and imported poultry meat. J Antimicrob Chemother 2009 Jul;64(1):207-209.
- 18. Bashir D, Thokar MA, Fomda BA, Bashir G, Zahoor D, Ahmad S, et al. Detection of metallo – beta – lactamase (MBL) producing Pseudomonas aeruginosa at a tertiary care hospital in Kashmir. Afr J Microbiol Res 2011;5(2):164-172.
- 19. Chakraborty D, Basu S, Das S. A study on infections caused by Metallo-Beta-Lactamase Producing Gram-negative Bacteria in Intensive Care Unit Patients. Am J Infect Dis 2010;6(2):34-39.
- 20. Leung GH, Gray TJ, Cheong EY, Haertsch P, Gottlieb T. Persistence of related bla-IMP-4 metallo-beta-lactamase producing Enterobacteriaceae from clinical and environmental specimens within a burns unit in Australia - a six-year retrospective study. Antimicrob Resist Infect Control 2013 Dec;2(1):35.
- 21. Aibinu I, Nwanneka T, Odugbemi T. Occurrence of ESBLs and MBL in Clinical Isolates of Pseudomonas aeruginosa from Lagos, Nigeria. J Am Sci 2007;3(4):81-85.
- 22. Akujobi CO, Odu NN, Okorondu SI. Detection of AmpC beta lactamases in clinical isolates of Escherichia coli and Klebsiella. Afr J Clin Exp Microbiol 2012;13(1):51-55.
- 23. Usha PT, Sabitha J, Nisha AR. Antimicrobial drug resistance: A global concern. Vet World 2010;3(3):138-139.
- 24. van den Bogaard AE, London N, Driessen C, Stobberingh EE. Antibiotic resistance of faecal Escherichia coli in poultry, poultry farmers and poultry slaughterers. J Antimicrob Chemother 2001 Jun;47(6):763-771.
- 25. Majalija S, Francis O, Sarah WG, Lubowa M. Antibiotic susceptibility profiles of fecal Escherichia coli isolates from dip-litter broiler chickens in Northern and Central Uganda. Vet Res 2010;3(4):75-80.
- 26. Ogunleye AO, Oyekunle MA, Sonibare AO. Multidrug resistant Escherichia coli isolates of poultry origin in Abeokuta, South Western Nigeria. Vet Arh 2008;78(6):501-509.
- 27. European Food Safety Authority (EFSA). EFSA panel on biological hazards; scientific opinion on the public health risks of bacterial strains producing extended-spectrum beta-lactamases and/or AmpC beta-lactamase in food and food-producing animals. EFSA J 2011;9(8):2322.
- 28. Manoharan A, Sugumar M, Kumar A, Jose H, Mathai D, Khilnani GC, et al; ICMR-ESBL group. Phenotypic & molecular characterization of AmpC β-lactamases among Escherichia coli, Klebsiella spp. & Enterobacter spp. from five Indian Medical Centers. Indian J Med Res 2012 Mar;135:359-364.
- 29. Mansouri S, Kalantar Neyestanaki D, Shokoohi M, Halimi S, Beigverdi R, Rezagholezadeh F, et al. Characterization of AmpC, CTX-M and MBLs types of β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli producing Extended Spectrum β-lactamases in Kerman, Iran. Jundishapur J Microbiol 2014 Feb;7(2):e8756.
- 30. Kolar M, Bardon J, Chroma M, Hricova K, Stosova T, Sauer P, et al. ESBL and AmpC beta-lactamase-producing Enterobacteriaceae in poultry in the Czech Republic. Vet Med (Praha) 2010;55(3):119-124.
- 31. Thomson KS. Extended-spectrum-β-lactamase, AmpC, and Carbapenemase issues. J Clin Microbiol 2010 Apr;48(4):1019-1025.
- 32. El-Hady SA, Adel LA. Occurrence and detection of AmpC β-lactamases among Enterobacteriaceae isolates from patients at Ain Shams University Hospital. Egypt J Med Hum Genet 2015;16:239-244.
- 33. Helmy MM, Wasfi R. Phenotypic and molecular characterization of plasmid mediated AmpC β-lactamases among Escherichia coli, Klebsiella spp., and Proteus mirabilis isolated from urinary tract infections in Egyptian hospitals. Biomed Research International, Hindawi Publishing Corporation 2014. p. 1-9.
- 34. Hussain M, Hasan F, Shah AA, Hameed A, Jung M, Rayamajhi N, et al. Prevalence of class A and AmpC β-lactamases in clinical Escherichia coli isolates from Pakistan Institute of Medical Science, Islamabad, Pakistan. Jpn J Infect Dis 2011;64(3):249-252.
- 35. Balkhair A. Antibiotics Resistance: When the Armamentarium Gets to the Verge of Being Empty. Oman Med J 2017 Jul;32(4):267-268.
- 36. Zorgani A, Almagatef A, Sufya N, Bashein A, Tubbal A. Detection of CTX-M-15 Among Uropathogenic Escherichia coli Isolated from Five Major Hospitals in Tripoli, Libya. Oman Med J 2017 Jul;32(4):322-327.