Although infections are caused by many pathogens, in high-risk hospital units like intensive care units (ICUs), research has shown that infections resulting from Staphylococcus aureus are on the rise emerging as a major public health concern to staff and patients in recent years.1,2 Methicillin-resistant S. aureus (MRSA) is a common nosocomial pathogen equipped with resistance and virulence factors which persist in hospitals and communities and can cause serious human and animal infections.3
Studies indicate a considerable proportion of resistance to many classes of antibiotics in clinical MRSA isolates.3–6 Many therapeutic options have been shown to be less effective for the treatment of MRSA infection because of the presence of multiresistant genes, which has resulted in clinical outcomes and increased morbidity and mortality rates.1,2 There is an apparent association between the pathogenesis of MRSA strains and the expression of virulence factors such as cell surface components (e.g., collagen-binding protein, clumping factor, fibronectin-binding protein, and elastin-binding protein) and secreted factors (e.g., staphylokinase, toxic shock syndrome toxin-1, hemolysin, Panton-Valentine leukocidin (PVL), exfoliative toxins (eta, etb), staphylococcal enterotoxins, and lipase).3
The development and spread of multidrug resistance (MDR) among MRSA isolates have been caused by the widespread use of beta-lactam, aminoglycoside, macrolides, and lincosamides over the last few decades.7,8 Aminoglycoside-modifying enzymes, including aminoglycoside phosphotransferase (APH), aminoglycoside acetyltransferase (AAC), and aminoglycoside nucleotidyltransferase, act as mediators in aminoglycoside resistance.5,6 Resistance to macrolides is associated with three mechanisms: i) target modification (mediated by erm genes); ii) efflux pumps (mediated by msr genes); and iii) enzymatic modification (mediated by lun genes). MupA and mupB genes act as mediators in mupirocin resistance. Research has shown that vancomycin is effective against S. aureus strain-related infections, but MRSA strains have increased alarmingly in recent years, which has reduced vancomycin susceptibility.9 VanA and vanB genes act as mediators in vancomycin resistance.9,10
Because of the increasing occurrence of S. aureus infections in ICU patients, awareness of the molecular and resistance patterns of this bacterium is imperative. Pulsed-field gel electrophoresis (PFGE), staphylococcal cassette chromosome mec (SCCmec) typing, accessory gene regulator (agr) typing, mec-associated hypervariable region (dru) typing, multilocus sequence typing (MLST), coagulase (coa) typing, and spa typing have been employed for typing S. aureus isolates.4,7,11–13 Although PFGE has been established as the standard method in recent years, it is believed that polymerase chain reaction (PCR)-based methods such as SCCmec and spa typing are ideal because of their cost-benefit, rapidity and high throughput capability.13 Three distinct regions (Fc, X, and C) are present in the spa gene. The polymorphic X region of the protein A gene has been sequenced by spa typing, which is a single-locus typing technique.8,13
Although the spa type distribution of S. aureus strains varies by geographic location, data from Iran indicates that spa types t790, t030, and t037 are widely disseminated in S. aureus clinical isolates.4,7,8,11 These also have been frequently reported in the Middle East, some European countries, and the United States.14,15
Resistance to methicillin is mediated by the mecA gene, which is carried within SCCmec. SCCmec elements are categorized according to differences in structure through SCCmec typing. Eleven types of SCCmec (I-XI) have been categorized through the mec gene and ccr gene complexes.16
With this background, it could be said that controlling staphylococcal infections is better achieved by identifying antibiotic resistance patterns, carriage of resistance genes, virulence determinants, and molecular types of S. aureus strains isolated from ICUs.1,9 We investigated the antibiotic resistance pattern, carriage of resistance and virulence determinants, and molecular typing of S. aureus isolates collected from ICUs based on SCCmec and spa genes.
Methods
Our cross-sectional research study was conducted between March and December 2017. Five university hospitals were designated to collect 84 non-duplicated MRSA strains isolated from clinical samples from ICU patients. These samples were obtained at the request of the specialist for medical reasons. The most common samples for MRSA strain isolation were wounds (n = 40; 47.6%), blood (n = 19; 22.6%), pus (n = 17; 20.2%), and catheters (n = 8; 9.5%). Out of 17 pus samples, six were drained from postoperative abscesses, four from breast abscesses, three from prostatic abscesses, two from a spinal epidural abscess, one from a lung abscess, and one from a salivary gland abscess. The Ethics Committee of Shahid Beheshti University of Medical Sciences in Tehran, Iran (IR.SBMU.SM.REC.1396.412) approved the research.
Preliminary identification was done by routine laboratory methods (colony morphology such as shape, size, color and hemolysis patterns, gram staining, tube coagulase tests, production of catalase tests, mannitol fermentation, and DNase plates). The isolates were confirmed to be S. aureus by detection of the femA and nucA genes using PCR.6
Mueller-Hinton agar (Merck; Germany) plates were used for phenotypic screening of methicillin resistance using cefoxitin 30 µg discs. As recommended by the Clinical and Laboratory Standard Institute (CLSI), 4% sodium chloride was added as a supplement.17 The criterion for choosing an isolate to investigate the presence of mecA gene was the observance of phenotypic methicillin resistance. After confirming the isolates as MRSA, they were stored in tryptic soy broth (Merck; Germany) for molecular testing along with 20% glycerol at -70 °C.
The susceptibility of each isolate was determined by the disk diffusion method as recommended by CLSI,17 except for ceftriaxone and fusidic acid, which were used according to the European Committee for Antimicrobial Susceptibility Testing (EUCAST) guidelines. Briefly, the inoculum was prepared from pure colonies of bacterial overnight culture, which were suspended in broth media or saline to an optical density equivalent to a 0.5 McFarland standard. Afterwards, Mueller-Hinton agar plates were seeded with S. aureus using a sterile cotton swab. Eventually, antibiotic disks were placed on the agar surface, and the plate was incubated at 35 °C overnight. After incubation, the diameter of the growth inhibition zone was measured in millimeters and interpreted using CLSI guidelines.17 The following antimicrobial agents (Mast; UK) were investigated: penicillin, ceftriaxone, kanamycin, ciprofloxacin, rifampicin, clindamycin, quinupristin-dalfopristin, tetracycline, erythromycin, linezolid, teicoplanin, fusidic acid, amikacin, tobramycin, gentamicin, and trimethoprim-sulfamethoxazole. The minimum inhibitory concentration (MIC) for vancomycin and mupirocin were determined by the broth microdilution test as recommended by CLSI.17 As instructed by CLSI, the broth microdilution test was performed to measure the MIC for vancomycin and mupirocin. MIC values of 8–256 µg/mL and 512 µg/mL were set to indicate low-level and high-level mupirocin resistance (LLMUPR, HLMUPR) of the strains, respectively.
As recommended by the EUCAST, fusidic acid MIC values of ≤ 1 µg/mL and > 1 µg/mL were set as signifying susceptible and resistant types, respectively. It is believed that, in addition to beta-lactams, MDR isolates cover resistance to three or more unique antibiotic classes.7,11,18 The standard strains for quality control were S. aureus ATCC25923 and ATCC29213.
The genomic DNA extraction for PCR assays was performed using the InstaGene matrix (BioRad; USA). This was done following manufacturer instructions with modifications such as the addition of 20 μL of 1 mg/mL lysostaphin (Sigma-Aldrich; USA). Isolates that exhibited phenotypic resistance to particular antimicrobial agents were tested for the presence of resistance genes (mecA, vanA, vanB, mupB, mupA, erm(A), erm(B), erm(C), msr(A), msr(B), tet(M), ant (4´)-Ia, aac (6´)-Ie/aph (2˝), aph (3´)-IIIa) using PCR.6,11,19 The carriage of genes that encode toxins such as etb, eta, pvl and tst, were examined using PCR as described.6,11
MRSA isolates underwent spa typing as recommended by Harmsen et al.13 Purified spa PCR products were subjected for DNA sequencing for both strands by Macrogen (Macrogen; South Korea). The sequences obtained were edited using Chromas software (version 1.45; Australia). Edited sequences were assigned to specific spa types using the Ridom Spa Server database (http://www.spaserver.ridom.de).
Multiplex-PCR was performed to type the SCCmec elements using specific primers as described by Boye et al.16 The following strains were used as references: ATCC 10442 (SCCmec type I), N315 (SCCmec type II), 85/2082 (SCCmec type III), MW2 (SCCmec type IV), and WIS (SCCmec type V).
Statistical analysis was performed using SPSS Statistics (SPSS Inc. Released 2009. PASW Statistics for Windows, Version 18.0. Chicago: SPSS Inc).

Figure 1: Lane M, 100-bp DNA Ladder (Fermentas, UK); Lane 1 nucA gene (270bp), Lane 2 eta gene (93bp), Lane 3 etb gene (276 bp), Lane 4 tst-1 gene (398 bp), Lane 5 luk-PV gene (180 bp), Lane 6 mecA gene (583 bp), Lane 7 tet(M) gene (158 bp), Lane 8 msr(A) gene (940 bp), Lane 9 msr(B) gene (595 bp), Lane 10 erm(A) gene (139 bp), Lane 11 erm(B) gene (141 bp), Lane 12 erm(C) gene (299 bp), Lane 13 ant(4΄)-Ia gene (135 bp), Lane 14 aph(3΄)-IIIa gene (269 bp), Lane 15 aac(6΄)-Ie/aph(2˝) gene (222 bp), and Lane 16 mupA gene (1158 bp).
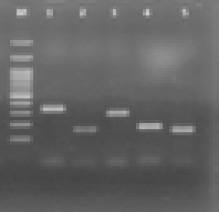
Figure 2: Lane M, 100-bp DNA ladder (Fermentas, UK); lanes 1–5, the variable polymerase chain reaction products of the spa gene.
Results
All isolates were MRSA. The number of MRSA isolates isolated from each hospital was as follows: 21 (25.0%) from hospital A, 18 (21.4%) from hospital B, 14 (16.7%) from hospital C, 12 (14.3%) from hospital D, and 19 (22.6%) from hospital E. Based on the results of the antimicrobial susceptibility testing, all 84 isolates were susceptible to vancomycin, linezolid, teicoplanin, and fusidic acid.
Higher resistance rates were observed for penicillin (72.6%) followed by gentamicin (59.5%), ceftriaxone (57.1%), kanamycin (48.8%), ciprofloxacin (46.4%), erythromycin (46.4%), tetracycline (42.9%), clindamycin (35.7%), rifampin (33.3%), mupirocin (26.2%), amikacin (20.2%), tobramycin (15.5%), and quinupristin-dalfopristin (14.3%). Resistance-associated genes demonstrated that 63.1% of isolates harbored the ant (4´)-Ia gene followed by aac (6´)-Ie/aph (2˝) (53.6%), aph (3´)-IIIa (35.7%), erm(A) (22.6%), tet(M) (15.5%), erm(B) (14.3%), msr(A) (13.1%), erm(C) (11.9%), msr(B) (9.5%), and mupA (9.5%). All isolates were negative for genes encoding vanA, vanB, and mupB. Among the 84 MRSA isolates investigated, the most frequent toxin gene was tst (25.0%). A low frequency of toxin genes was noted in isolates with pvl (14.3%), eta (3.6%), and etb (2.4%) genes [Figure 1].
Of the 84 MRSA isolates, 78.6% (66/84) were defined as MDR. The predominant simultaneous resistance patterns were for six antibiotics (17.9%, 15/84) followed by three antibiotics (13.1%, 11/84), seven antibiotics (11.9%, 10/84), nine antibiotics (10.7%, 9/84), and five antibiotics (6.0%, 5/84). Of the 22 mupirocin-resistant MRSA isolates, eight (9.5%) had high resistance levels, and 14 (16.7%) had low resistance levels. All the HLMUPR strains were collected from wound samples. Constitutive and inducible phenotypes were detected in 30 (35.7%) and 9 (10.7%) MRSA isolates, respectively.
SCCmec typing showed that type III was predominant and present in 50 isolates (59.5%), followed by type IV in 34 isolates (40.5%). The spa typing discriminated seven types: t388 (36.9%), t852 (14.3%), t924 (13.1%), t790 (11.9%), t064 (10.7%), t037 (9.5%), and t084 (3.6%). Of the 12 isolates carrying the pvl gene, eight (66.7%) belonged to t852 and four (33.3%) belonged to t790. All the HLMUPR-MRSA strains belonged to the t064 spa type while isolates with a LLMUPR belonged to t790 (10 isolates), t084 (three isolates) and t064 (nine isolate) spa types.
The findings revealed that spa types circulating in hospital A were t388 (11 isolates), t852 (four isolates), and t064, t790 and t084 (two isolates each). Those circulating in hospital B were t388 and t037 (five isolates each), t790 and t924 (three isolates each), and t064 (two isolates) [Figure 2]. Circulating clones in hospital C were t924 (six isolates), t852 and t790 (three isolates each), t388 and t064 (one isolate each). Circulating clones in hospital D were t388 (six isolates), t852 (five isolates), and t084 (one isolate). The spa types detected in hospital E were t388 (eight isolates), t064 (four isolates), t037 (three isolates), t790 (two isolates), and t924 (two isolates). These spa types showed wide differences in the carriage of resistance and toxin genes. The characteristics related to the 84 MRSA isolates are presented in Table 1.
Table 1: Molecular characterization of MRSA strains isolated from ICUs.
|
t388 |
III |
tst (12, 38.7) |
mecA (31, 100), ant(4´)-Ia (25, 80.6), aac(6´)-Ie/aph(2˝)(18, 58.1), aph(3´)-IIIa (14, 45.2),erm(A) (9, 29.0) |
GM (6, 19.4) |
31 (36.9) |
| |
|
|
|
PG, CRO, RI, E, CD, T, AK, GM (10, 32.3) |
|
| |
|
|
|
K, RI (2, 6.4) |
|
| |
|
|
|
PG, CIP, K, E, CD, GM (7, 22.6) |
|
| |
|
|
|
PG, CRO, CIP (6, 19.4) |
|
|
t852 |
IV |
pvl (8, 66.7) |
mecA (12, 100), ant(4´)-Ia (8, 66.7), erm(B) (7, 58.3), msr(A) (5, 41.7) |
CRO, K, RI, T, AK (3, 25.0) |
12 (14.3) |
| |
|
|
|
PG, CIP, K, E, CD, GM (4, 33.3) |
|
| |
|
|
|
GM (5, 41.7) |
|
|
t924 |
III |
tst (4, 36.4) |
mecA (11, 100), aac(6´)-Ie/aph(2˝) (7, 63.6), aph(3´)-IIIa (6, 54.5), erm(A) (7, 63.6), erm(C) (4, 36.4), msr(B) (5, 45.5), tet(M) (3;27.3) |
PG, CRO, RI, E, CD, T, AK, GM (2, 18.2) |
11 (13.1) |
| |
|
|
|
CRO, K, RI, T, AK (1, 9.1) |
|
| |
|
|
|
PG, CIP, K, E, CD, GM (4, 36.4) |
|
| |
|
|
|
PG, CRO, CIP (3, 27.3) |
|
| |
|
|
|
PG, CRO (1, 9.1) |
|
|
t790 |
IV |
pvl (4, 40.0), eta (1, 10.0) |
mecA (10, 100), ant(4´)-Ia (10, 100), aac(6´)-Ie/aph(2˝) (9, 90.0), aph(3´)-IIIa (7, 70.0), tet(M) (6, 60.0) |
PG, CRO, K, CIP, T, TN, MUP (6, 60.0) |
10 (11.9) |
| |
|
|
|
PG, CRO, K, RI, T, SYN, MUP, E, GM (4, 40.0) |
|
|
t064 |
IV |
|
mecA (9, 100), ant(4´)-Ia (9, 100), aac(6´)-Ie/aph(2˝) (9, 100), erm(C) (6, 66.7), msr(A) (6, 66.7), tet(M) (4, 44.4), mupA (8, 88.9) |
PG, CRO, K, CIP, T, TN, MUP (4, 44.4) |
9 (10.7) |
| |
|
|
|
PG, CRO, K, RI, T, SYN, MUP, E, GM (5, 55.6) |
|
|
t037 |
III |
tst (5, 62.5) |
mecA (8, 100), ant(4´)-Ia (1, 12.5), erm(B) (3, 37.5), msr(B) (3, 37.5) |
PG, CRO, CIP (2, 25.0) |
8 (9.5) |
| |
|
|
|
CRO, K, RI, T, AK (1, 12.5) |
|
| |
|
|
|
No resistance (5, 62.5) |
|
|
t084 |
IV |
eta (2, 66.7), etb (2, 66.7) |
mecA (3, 100), aac(6´)-Ie/aph(2˝) (2, 66.7), aph(3´)-IIIa (3, 100), erm(A) (3, 100), erm(B) (2, 66.7) |
PG, CIP, CD, E, SYN, MUP, TN, GM (2, 66.7) |
3 (3.6) |
MRSA: methicillin-resistant S. aureus; ICUs: intensive care units; PG: penicillin; CRO: ceftriaxone; CD: clindamycin; E: erythromycin; GM: gentamicin;
RI: rifampin; T: tetracycline; CIP: ciprofloxacin; K: kanamycin; SYN: quinupristin-dalfopristin; AK: amikacin; TN: tobramycin; MUP: mupirocin.
Discussion
We made several important findings in relation to spa typing of MRSA strains isolated from ICUs, including detection of t388 as the predominant spa type and a relatively high resistance to mupirocin (26.2%), which was distributed among spa types t064 (nine isolates), t790 (10 isolates), and t084 (three isolates). The distribution of the spa types varies from one geographical region to another.5,8,20 In contrast to our finding that t388 was the predominant type of MRSA isolated from ICUs (36.9%), another study from Iran reported a low frequency of t388 (4.4%) in MRSA strains isolated from burn patients.21 A study from Taiwan also showed that among 157 MRSA strains isolated from blood cultures in nine medical centers, only one isolate was t388.22 It appears that the prevalence of this spa type has increased significantly over time and was successfully established as the dominate spa type among MRSA isolates in the hospital studied. In our study, none of the t388 strains harbored pvl genes, and fewer than half of isolates (38.7%) carried the tst gene. PVL-negative and tst-positive t388 strains have been reported by several investigators.21,22
The spa type t852 was the second most common type among the tested isolates (14.3%). More than half of the t852 isolates carried pvl genes (66.7%). PVL-positive t852 strains have been described in Oman,23 Saudi Arabia,24 Qatar,25 and some European countries,14,15 suggesting an increase in the transmission of this variant Persian Gulf countries.
We detected spa type t924 in 13.1% of tested MRSA isolates. It was also found that all t924 isolates belonged to SCCmec III. Similarly, a study from Poland published in 2018 of 215 isolates obtained from the airways of 107 patients with cystic fibrosis documented a low prevalence of t924.7 In our study, analysis of resistance genes showed that 27.2% of isolates were resistant to rifampin. A multicenter study replicated these findings.26
In contrast to studies which indicated SCCmec IV/t790 as the most common spa type in Iran,27 our study found a prevalence of this spa type in 11.9% of isolates. The low frequency of t790 in our study is in line with the results of studies from Kuwait14 and Iran.11 As in the results of previous studies from other countries, spa type t790 was found in both PVL-negative and -positive MRSA strains.28 It was also found that most spa t790 isolates belonged to SCCmec IV, which is in agreement with a previous study from Kuwait.14 All t790 MRSA isolates were resistant to mupirocin and demonstrated the LLMUPR phenotype and were positive for many antibiotic resistant genes, including mecA (10, 100%), ant(4´)-Ia (10, 100%), aac(6´)-Ie/aph(2˝) (9, 90%), aph(3´)-IIIa (7, 70%) and tet(M) (6, 60%). This finding is in accordance with a study from Ireland, which showed the presence of aminoglycoside resistant genes in the majority of their isolates.29
The frequency of the SCCmec IV/t064 strain was 10.7% in our study. Eight of nine (88.9%) MRSA strains were found to carry the mupA gene. One study of the clonal distribution of MRSA in Kuwait over 12 years in 13 public hospitals reported the t064 spa type with an HLMUPR pattern that carried different resistance genes.14 In line with the results of our study, this spa type also has been reported in Nigeria30 and Ireland.15
Another study from Iran reported multiresistant SCCmec III/t037 MRSA as the major spa type circulating in hospitalized patients in ICUs.11 We found that only 9.5% of isolates belonged to the t037 spa type. Our data showed that the t037 strains were PVL-negative, but were positive for the tst gene in addition to mecA (8; 100%), ant(4´)-Ia (1; 12.5%), erm(B) (3; 37.5%), and msr(B) (3; 37.5%). This spa type has been reported in Saudi Arabia, China, Iran and among hospital acquired-MRSA isolates in Europe, the United States, and other regions of Asia.14,15,27 Although t037 MRSA isolates harboring the vanA gene have been detected by several investigators,10,31,32 our study showed that none of the t037 isolates were resistant to vancomycin.
The t084 spa type was identified among the tested MRSA strains. Our study results indicate that all SCCmec IV/t084 strains were mupirocin resistant. This is consistent with another Iranian study, which detected SCCmec IV/t084 (40%) strains among hospital- and community acquired-MRSA isolates.18
Published evidence suggests that SCCmec types I, II, and III are common among hospital acquired-MRSA isolates; however, the significant features of community acquired-MRSA are SCCmec types IV and V.11,16 These can act as epidemiological markers for hospital- and community acquired-MRSA differentiation.
The results of SCCmec typing disclosed that type III was the predominant SCCmec type (59.5%) associated with MDR pattern among the MRSA isolates consistent with Japoni-Nejad et al.33 A wide distribution of SCCmec type III among the isolates in the current study emphasizes the nosocomial origin of the samples. This is indicated by the broad distribution of SCCmec type III among our isolates.
The frequency of erm(A), erm(B), msr(A), erm(C), and msr(B) genes was 22.6%, 14.3%, 13.1%, 11.9%, and 9.5%, respectively. This finding is in line with Nezhad et al,6 who reported the erm(A) gene as the predominant macrolide resistance gene among MRSA strains isolated from hospitalized patients in ICUs.
Aminoglycoside resistance represents a challenge for curing serious staphylococcal infections.5 The most significant gene identified among the isolates of the current study was the ant(4´)-Ia gene, followed by (6´)-Ie/aph (2˝) (53.6%), and aph(3´)-IIIa (35.7%). This is consistent with previous studies.6,34 Although another study demonstrated that tet(M) is the most common antibiotic resistance gene detected in MRSA strains (54.05%).35 The results of our study show a low resistance rate for tet(M) (15.5%) in the isolates.
Conclusion
We recorded a high frequency of resistance among the specific spa types. Therefore, identification and screening of dominant spa types and SCCmec elements could aim to evaluate the significance of distribution of these MRSA isolates and their clonal relationships and allow selection of proper treatment protocols and successful implementation of antibiotic stewardship programs.
Disclosure
The authors declared no conflicts of interest. This study was supported financially by the grant (No. 12203) from Research Deputy of Shahid Beheshti University of Medical Sciences.
references
- 1. Lucet J-C, Chevret S, Durand-Zaleski I, Chastang C, Régnier B; Multicenter Study Group. Prevalence and risk factors for carriage of methicillin-resistant Staphylococcus aureus at admission to the intensive care unit: results of a multicenter study. Arch Intern Med 2003 Jan;163(2):181-188.
- 2. Klevens RM, Edwards JR, Tenover FC, McDonald LC, Horan T, Gaynes R; National Nosocomial Infections Surveillance System. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus in intensive care units in US hospitals, 1992-2003. Clin Infect Dis 2006 Feb;42(3):389-391.
- 3. Gordon RJ, Lowy FD. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis 2008;46(Suppl 5):S350-S359.
- 4. Mashouf RY, Hosseini SM, Mousavi SM, Arabestani MR. Prevalence of enterotoxin genes and antibacterial susceptibility pattern of Staphylococcus aureus strains isolated from animal originated foods in West of Iran. Oman Med J 2015 Jul;30(4):283-290.
- 5. Asadollahi P, Farahani NN, Mirzaii M, Khoramrooz SS, van Belkum A, Asadollahi K, et al. Distribution of the most prevalent spa types among clinical isolates of methicillin-resistant and-susceptible Staphylococcus aureus around the world: A review. Front Microbiol 2018 Feb;9:163.
- 6. Nezhad RR, Meybodi SM, Rezaee R, Goudarzi M, Fazeli M. Molecular characterization and resistance profile of methicillin resistant Staphylococcus aureus strains isolated from hospitalized patients in intensive care unit, Tehran-Iran. Jundishapur J Microbiol 2017;10(3):e41666.
- Garbacz K, Piechowicz L, Podkowik M, Mroczkowska A, Empel J, Bania J. emergence and spread of worldwide Staphylococcus aureus clones among cystic fibrosis patients. Infect Drug Resist. 2018;11:247.
- 8. Taheri N, Ardebili A, Amouzandeh-Nobaveh A, Ghaznavi-Rad E. Frequency of antiseptic resistance among Staphylococcus aureus and coagulase-negative staphylococci isolated from a university hospital in central Iran. Oman Med J 2016 Nov;31(6):426-432.
- 9. Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest 2003 May;111(9):1265-1273.
- 10. Appelbaum PC, Bozdogan B. Vancomycin resistance in Staphylococcus aureus. Clin Lab Med 2004 Jun;24(2):381-402.
- 11. Goudarzi M, Goudarzi H, Sá Figueiredo AM, Udo EE, Fazeli M, Asadzadeh M, et al. Molecular characterization of methicillin resistant Staphylococcus aureus strains isolated from intensive care units in Iran: ST22-SCCmec IV/t790 emerges as the major clone. PLoS One 2016 May;11(5):e0155529.
- 12. Zinn CS, Westh H, Rosdahl VT, Group SS; Sarisa Study Group. An international multicenter study of antimicrobial resistance and typing of hospital Staphylococcus aureus isolates from 21 laboratories in 19 countries or states. Microb Drug Resist 2004;10(2):160-168.
- 13. Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol 2003 Dec;41(12):5442-5448.
- 14. Boswihi SS, Udo EE, Al-Sweih N. Shifts in the clonal distribution of methicillin-resistant Staphylococcus aureus in Kuwait hospitals: 1992-2010. PLoS One 2016 Sep;11(9):e0162744.
- Shore A, Rossney AS, Keane CT, Enright MC, Coleman DC. Seven novel variants of the staphylococcal chromosomal cassette mec in methicillin-resistant Staphylococcus aureus isolates from Ireland. Antimicrob Agents Chemother. 2005;49:2070–83.
- 16. Boye K, Bartels MD, Andersen IS, Møller JA, Westh H. A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I-V. Clin Microbiol Infect 2007 Jul;13(7):725-727.
- 17. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. 27th ed. CLSI supplement M100.
- 18. Fasihi Y, Kiaei S, Kalantar-Neyestanaki D. Characterization of SCCmec and spa types of methicillin-resistant Staphylococcus aureus isolates from health-care and community-acquired infections in Kerman, Iran. J Epidemiol Glob Health 2017 Dec;7(4):263-267.
- 19. Udo EE, Jacob LE, Mathew B. Genetic analysis of methicillin-resistant Staphylococcus aureus expressing high- and low-level mupirocin resistance. J Med Microbiol 2001 Oct;50(10):909-915.
- 20. Shakeri F, Shojai A, Golalipour M, Rahimi Alang S, Vaez H, Ghaemi EA. Spa diversity among MRSA and MSSA strains of staphylococcus aureus in north of Iran. Int J Microbiol 2010;2010:351397.
- 21. Ohadian Moghadam S, Pourmand MR, Mahmoudi M, Sadighian H. Molecular characterization of methicillin-resistant Staphylococcus aureus: characterization of major clones and emergence of epidemic clones of sequence type (ST) 36 and ST 121 in Tehran, Iran. FFEMS Microbiol Lett 2015;362(8):fnv043.
- 22. Ho C-M, Ho M-W, Li C-Y, Lu J-J. Fine typing of methicillin-resistant Staphylococcus aureus isolates using direct repeat unit and staphylococcal interspersed repeat unit typing methods. J Microbiol Immunol Infect 2015 Aug;48(4):370-375.
- 23. Udo EE, Al-Lawati BA, Al-Muharmi Z, Thukral SS. Genotyping of methicillin-resistant Staphylococcus aureus in the Sultan Qaboos University Hospital, Oman reveals the dominance of Panton-Valentine leucocidin-negative ST6-IV/t304 clone. New Microbes New Infect 2014 Jul;2(4):100-105.
- 24. Monecke S, Skakni L, Hasan R, Ruppelt A, Ghazal SS, Hakawi A, et al. Characterisation of MRSA strains isolated from patients in a hospital in Riyadh, Kingdom of Saudi Arabia. BMC Microbiol 2012 Jul;12(1):146.
- 25. El-Mahdy T, El-Ahmady M, Goering R. Molecular characterization of methicillin-resistant Staphylococcus aureus isolated over a 2-year period in a Qatari hospital from multinational patients. Clin Microbiol Infect 2014;20(2):169-173.
- 26. Wichelhaus T, Schäfer V, Brade V, Böddinghaus B. Differential effect of rpoB mutations on antibacterial activities of rifampicin and KRM-1648 against Staphylococcus aureus. J Antimicrob Chemother 2001 Feb;47(2):153-156.
- 27. Alreshidi MA, Alsalamah AA, Hamat RA, Neela V, Alshrari AS, Atshan SS, et al. Genetic variation among methicillin-resistant Staphylococcus aureus isolates from cancer patients in Saudi Arabia. Eur J Clin Microbiol Infect Dis 2013 Jun;32(6):755-761.
- 28. Shore AC, Rossney AS, Brennan OM, Kinnevey PM, Humphreys H, Sullivan DJ, et al. Characterization of a novel arginine catabolic mobile element (ACME) and staphylococcal chromosomal cassette mec composite island with significant homology to Staphylococcus epidermidis ACME type II in methicillin-resistant Staphylococcus aureus genotype ST22-MRSA-IV. Antimicrob Agents Chemother 2011 May;55(5):1896-1905.
- 29. Shore AC, Tecklenborg SC, Brennan GI, Ehricht R, Monecke S, Coleman DC. Panton-Valentine leukocidin-positive Staphylococcus aureus in Ireland from 2002 to 2011: 21 clones, frequent importation of clones, temporal shifts of predominant methicillin-resistant S. aureus clones, and increasing multiresistance. J Clin Microbiol 2014 Mar;52(3):859-870.
- 30. O’Malley SM, Emele FE, Nwaokorie FO, Idika N, Umeizudike AK, Emeka-Nwabunnia I, et al. Molecular typing of antibiotic-resistant Staphylococcus aureus in Nigeria. J Infect Public Health 2015 Mar-Apr;8(2):187-193.
- 31. Azimian A, Havaei SA, Fazeli H, Naderi M, Ghazvini K, Samiee SM, et al. Genetic characterization of a vancomycin-resistant Staphylococcus aureus isolate from the respiratory tract of a patient in a university hospital in northeastern Iran. J Clin Microbiol 2012 Nov;50(11):3581-3585.
- 32. Havaei SA, Azimian A, Fazeli H, Naderi M, Ghazvini K, Samiee SM, et al. Genetic characterization of methicillin resistant and sensitive, vancomycin intermediate Staphylococcus aureus strains isolated from different Iranian Hospitals. ISRN Microbiol 2012;2012:215275.
- 33. Japoni-Nejad A, Rezazadeh M, Kazemian H, Fardmousavi N, van Belkum A, Ghaznavi-Rad E. Molecular characterization of the first community-acquired methicillin-resistant Staphylococcus aureus strains from Central Iran. Int J Infect Dis 2013 Nov;17(11):e949-e954.
- 34. Rahimi F. Characterization of resistance to aminoglycosides in methicillin-resistant staphylococcus aureus strains isolated from a tertiary care hospital in Tehran, Iran. Jundishapur J Microbiol 2016 Jan;9(1):e29237.
- 35. Dormanesh B, Siroosbakhat S, Khodaverdi Darian E, Afsharkhas L. Methicillin-resistant Staphylococcus aureus isolated from various types of hospital infections in pediatrics: Panton-Valentine leukocidin, staphylococcal chromosomal cassette mec SCCmec phenotypes and antibiotic resistance properties. Jundishapur J Microbiol 2015 Nov;8(11):e11341.