Urinary tract infections (UTIs) are the most prevalent bacterial infections in humans, and Escherichia coli is the most important cause of nosocomial UTIs. Narrow- and extended-spectrum cephalosporins have been used to treat such infections.1 Management of UTIs has become increasingly problematic owing to the increasing production of extended-spectrum β-lactamases (ESBLs).2 The enzymes TEM and SHV were previously recognized as the main ESBLs; however, CTX-M has recently become more prominent and is considered to be the most prevalent β-lactamases found in clinical isolates of E. coli globally.3 A nationwide survey in the United States during 2009–2010 found 91% of ESBL-producing E. coli strains carried CTX-M-type genes.4 CTX-M-type ESBLs are also dominant in European and Asian countries.3,5 Similarly, it has been reported from many African countries like Egypt, Algeria, Tunisia, Tanzania, and Cameroon.6–10 Within the CTX-M family, CTX-M-15 is currently the most widely disseminated genotype associated with major causes of infections in both community and hospital environments.11,12 The wide dissemination of CTX-M-15 producing E. coli isolates has been explained by many workers. First, it has been proposed that the strain virulence background could be involved in this dissemination process. In fact, many reports have shown that CTX-M-15 is closely associated with the international and pandemic uropathogenic ST131 clone, which have specific virulence factors.4,13 Second, the association of CTX-M-15 with the IncF plasmids, which are well adapted to E. coli, may facilitate the spread of this determinant in the E. coli population.14 In addition to virulence background and IncF plasmids bearing CTXM-15, it was recently suggested that the association of various plasmid addiction systems might contribute to plasmid maintenance in their host.15–17 An addiction system or a toxin-antitoxin system helps maintain plasmids in bacteria during host replication by killing of plasmid-free cells resulting from segregation or replication defects.18
To date, few studies have been carried out in Libya to establish the associations between the clinical origin of strains, resistance-encoding phenotypes, and phylogenetic groups among E. coli isolates.19,20 The aim of the present investigation was to determine the prevalence of E. coli isolates associated with UTIs in patients attending five hospitals, their susceptibility to antimicrobial agents, and the presence of CTX-M-15 among ESBL producing isolates of E. coli by polymerase chain reaction (PCR).
Methods
A total of 346 urine samples were collected from hospitalized patients with UTIs from five teaching hospitals in Tripoli: Tripoli Medical Centre (TMC), Tripoli Pediatric Hospital (TPH), Khadra Hospital, Aljala Hospital, and Tripoli Central Hospital (TCH). All urine samples were taken as part of the clinical work-up and these were included in this prospective laboratory-based surveillance study. The study was carried out during 2013 (January–December).
In this investigation, urine specimens were collected under approved ethical standards and the study was reviewed and approved by the faculty of Pharmacy, University of Tripoli, and participating hospitals. Urine specimens were obtained from all patients with supervision of a medical doctor or senior nurse for patients aged one year or older; midstream urine was collected in a sterile container and processed in the laboratory within two hours of collection. Urine specimens were obtained by suprapubic aspiration from children who were not toilet trained and processed as above. The consent of the patient or their guardian was obtained before urine specimens were collected. Using calibrated loops, urine specimens were inoculated on to MacConkey and blood agar plates. After 18–24 hour incubation at 37 oC, the number of colony forming units (cfu) was counted, and urine samples giving ≥ 105 cfu/mL of urine were considered significant.
All specimens were cultured on different media by standard bacteriological procedures. Isolated organisms were identified at the species level and tested for their susceptibility to a variety of antimicrobial agents using the BD Phoenix Automated Microbiology System (PAMS, MSBD Biosciences, Sparks Md, USA) according to the manufacturer’s instructions. The system uses combination panels for identification (ID) and antimicrobial susceptibility testing (AST) of bacteria. These include the Phoenix NMIC/ID Panels intended for in vitro rapid ID and AST by minimal inhibitory concentration (MIC) of Gram-negative aerobic and facultative anaerobic bacteria from pure culture belonging to the Enterobacteriaceae and non-Enterobacteriaceae families. Confirmatory tests for ESBL production were performed with all of the isolates initially identified by the Phoenix system. Phenotypic confirmation of ESBLs was done using E-test strips containing ceftazidime, and ceftazidime-clavulanate was used to determine the MIC ratio according to the manufacturer’s instructions (epsilometer assay; AB Biodisk Slona, Sweden) performed on Mueller-Hinton agar. All isolates were screened and interpreted for ESBL phenotype according to the criteria of the Clinical and Laboratory Standards Institute.21 Multidrug-resistant (MDR) bacteria were defined as showing resistance to three or more different classes of antibiotics such as fluoroquinolones, aminoglycosides, and cephalosporins.22 Reference strain of E. coli ATCC 25922, E. coli ATCC 35218, and Klebsiella pneumoniae ATCC 700603 were used as controls.
ESBL producing E. coli isolates were screened for the blaCTX-M-15 gene using previously reported primers.23 The plasmids were isolated using the QIAGEN Plasmid Mini Kit according to the manufacturer’s instructions. The reaction mixture contained a total of 25 µl: 5 µl of 5X Red Load Taq Mix composed of Taq Polymerase, 0.05 u/µl dNTPs (200 µM) (dATP, dCTP, dGTP, dTTP) reaction buffer with KCl and MgCl2 (1.5 mM) red dye, gel loading buffer, stabilizers (Metabion, Martinsried- Germany); 0.5 µl of each primer 10 pmol/µl; and extracted plasmid DNA (2–50 ng). The thermal profile included one cycle of initial denaturation at 95 oC for 2 minutes followed by 35 cycles at 95 oC for 30 seconds, annealing at 52 oC for 30 seconds, and extensions at 72 oC for 45 seconds. The PCR reaction was carried out with a TC-412 thermocycler (Techne, Duxford, Cambridge): 5 µl of the PCR amplification products were electrophoresed in agarose (2% m/v) containing 0.5 ug/mL ethidium bromide. The amplified PCR products were visualized under UV light and electronically documented (MultiDoc-It Digital Imaging System UVP, Cambridge, UK). A 50 bp DNA ladder (Metabion, Martinsried- Germany) was used as a molecular size marker.
All statistical analyses were performed with the SPSS Statistics (SPSS Inc. Released 2007. SPSS for Windows, Version 16.0. Chicago, SPSS Inc.). Statistical analyses were performed using the chi-square test. The statistical significance was set at p = 0.050.
Results
A total of 346 isolates were collected and confirmed as uropathogenic E. coli by the Phoenix automated system. The age of the patients with UTI ranged from < 1–90 years old. The distribution of ESBL-producing E. coli varied among the different hospitals of Tripoli. The majority were isolated from patients at TMC (67.6%) compared with other hospitals; TPH (9.9%), Khadra Hospital (9.9%), Aljala Hospital (8.4), and TCH (4.2%). The majority of the isolates expressing ESBL were obtained from female patients 51/71 (71.8%) and 20/71 (28.2%) were obtained from males.
The results of antimicrobial susceptibility tests for ESBL-producing 71/346 (20.5%) strains and non-ESBL producers 275/346 (79.5%) are summarized in Table 1. Extremely high resistance rates (p < 0.001) were observed for ceftriaxone, cefepime, and ceftazidime (93.0–100.0%) among ESBL producers compared to non-ESBL producers (2.2–4.7%). High rates of resistance were also demonstrated to ciprofloxacin (67.6%) and cefotaxime (64.8%). Fewer isolates (2.8%) were resistant to amikacin, meropenem, and ertapenem. ESBL producers were more often resistant to major classes of antibiotics compared with non-ESBl producers. MDR was documented for 77/346 (22.2%) of isolates, and was significantly higher (p < 0.001) among ESBLs compared with non-ESBL producer uropathogenic E. coli.
All 71 ESBL-positive E. coli isolates were confirmed by E-test. PCR analysis found 61/346 (17.6%) of isolates contained blaCTX-M-15, the majority of these isolates 61/71 (85.9%) that harbored CTX-M-15 gene were found to be ESBL producers [Figure 1]. There was no significant difference between gender and expression of CTX-M-15 gene; however, there was a correlation between expression of CTX-M-15 and resistance to ceftazidime (p < 0.001, odd ratio 179.142 [59.2–541.9]). CTX-M-15-positive isolates were highly sensitive to ertapenem, meropenem, and amikacin (97.2%). It was observed that the majority of isolates 44/71 (61.9%) defined as MDR organisms also had blaCTX-M-15 gene and classified as ESBL positive.
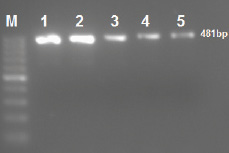
Figure 1: Amplification of CTX-M-15 gene in extended-spectrum β-lactamase producing Escherichia coli isolates by single polymerase chain reaction. Lane M 50 bp ladder; Lanes 1 to 5 CTX-M-15-producing isolates. The amplified amplicon size is 481 bp.
Table 1: Antimicrobial resistance patterns of Escherichia coli from patients with urinary tract infections.
|
Ampicillin |
71 (100.0) |
196 (71.2) |
0.004 |
|
Amikacin |
2 (2.8) |
4 (1.4) |
0.440 |
|
Gentamicin |
27 (38.0) |
32 (11.6) |
< 0.001 |
|
Cefoxitin |
27 (38.0) |
22 (8.0) |
< 0.001 |
|
Ceftriaxone |
69 (97.1) |
13 (4.7) |
< 0.001 |
|
Cefepime |
71 (100.0) |
6 (2.2) |
< 0.001 |
|
Ceftazidime |
66 (93.0) |
11 (4.0) |
< 0.001 |
|
Nitrofurantoin |
12 (16.9) |
15 (5.4) |
0.002 |
|
Meropenem |
2 (2.8) |
0 (0.0) |
0.055 |
|
Ciprofloxacin |
48 (67.6) |
91 (33.1) |
< 0.001 |
|
Cefotaxime |
46 (64.8) |
112 (40.7) |
0.0004 |
|
Ertapenem |
2 (2.8) |
0 (0.0) |
0.055 |
*n = 77. ESBL: extended-spectrum β-lactamase; MDR: Multidrug resistance.
Discussion
This study provides molecular-epidemiological data on ESBL E. coli isolated from patients in the five university hospitals of Tripoli in Libya during 2013. The distribution of ESBL-producing E. coli varied (4.2–67.6%) among the different hospitals of Tripoli; the highest proportions (67.6%) was observed among isolates obtained from the largest tertiary teaching hospital, TMC. In this study, ESBL was detected in a higher proportion of isolates (20.5%) compared to a previous study that found ESBL in 6.7% of uropathogenic E. coli.11 These results were consistent with the results of reports from the Middle East.24,25
Identification of ESBL among isolates varied from 66.7% in India,26 54.7–61% in Turkey, and 72.1% in Iran.26–28 These figures were lower in America and Europe.3,27 SMART data have shown that the Middle East has the second highest ESBL prevalence after Asia (around 37% for UTI isolates).29 The Middle East is the only region in the world where the prevalence of ESBLs has been increasing significantly for UTIs,29 ESBL in UTI-associated E. coli isolates from Lebanese patients, increased consistently from 2.3% in 2000 to 16.8% in 2009.30 Three studies carried out in Saudi Arabia revealed that an increasing prevalence of ESBL E. coli isolated from UTI over two years, 2013 = 20.4%, 2014 = 33.3%, and 2014 = 35.8%.24,31,32 Although the prevalence of ESBLs-producing bacteria is a worldwide problem, identification of these isolates can vary by countries and institutions within a country.33
The findings of the present study identified CTX-M-15 among E. coli isolated from UTI in Tripoli, Libya. The results indicated that overall 17.3% of clinical E. coli isolated carried CTX-M-15 gene, the majority (85.9%) of ESBL-carrying E. coli isolates CTX-M-15 gene. The results of this study are consistent with the global distribution of CTX-M-15.5,34 It is worth noting that although some of the CTX-M enzymes have been associated with specific countries, such as CTX-M-9 and CTX-M-14 in Spain, CTX-M-1 in Italy, and CTX-M-2 in Israel, Japan, and most South American countries.35,36 Others, such as CTX-M-15, have been detected worldwide.5,12,35,36 In Arab countries, the first description of CTX-M was in Egypt and then in the United Arab Emirates and Kuwait.37–39 In these countries, the predominant ESBL was CTX-M-15. After 2000, the prevalence of CTX-M increased steadily in the Tunisian healthcare setting especially CTX-M-15, which in many studies was most prevalent among ESBLs.40,41 Mnif et al,42 reported that 72% of ESBL-positive E. coli isolates were CTX-M producers. High rates were demonstrated in Saudi Arabia; CTX-M-type enzymes accounted for 100% of all ESBLs, and the predominant ESBL was CTX-M-15 (92.1%). A similar trend (98%) was observed in Iran.43 SMART study (2011–2013) conducted in Lebanon and Jordan showed that CTX-M-15 was the most prevalent ESBL produced.44 This results are similar to those for Lebanon; the majority of the strains (83%) express CTX-M-15 ESBL.45 Previously, Abujnah and colleagues in Libya reported that 6/13 of E. coli isolates expressed blaCTX-M gene, but they failed to determine the prevalence of CTX-M-15.19
Xia et al,46 reported low rates (18.2%) of blaCTX-M-15 ESBL-producing E. coli isolated from patients with community-onset or hospital-onset infections in China. Detection of CTX-M-15 among all studied isolates (17.3% overall) was lower than Argentina where the occurrence of CTX-M-15 was 45.2% among E. coli isolates.47 Similarly, in Lithuania the most common CTX-M β-lactamase among the E. coli isolates, collected during a period of five years, was CTX-M-15 followed by CTX-M-14 and CTX-M-92 (56.7%, 13.3%, and 11% respectively).48 These results reflect the global trend toward a pandemic spread of CTX-M-type ESBLs in E. coli.34
The prevalence and spread of antibiotic resistance and MDR E. coli isolates was previously determined.19 Extremely high resistance rates were observed to ceftriaxone, cefepime, and ceftazidime among ESBL producers (93.0–100.0%) compared to non-ESBL producers (2.2–4.7%). High rates of resistance were also demonstrated to ciprofloxacin (67.7%) and cefotaxime (64.8%). It is very important to emphasize that carbapenems maintained excellent activity (97.8%) against ESBL-producing E. coli isolates during the study period. Overall, MDR were detected in 22.2% of isolates lower than previously detected in 32.2% of E. coli isolated from patients with UTIs attending Zawia Hospital, in Libya.24 The majority (62%) of isolates simultaneously defined to be MDR organisms were also found to express the blaCTX-M-15 gene and were designated ESBL positive. Alzohairy and Khadri reported higher rates of MDR among E. coli (53.6%) isolated from community and hospital-acquired UTIs in Qassim province, Saudi Arabia.49 Generally, most CTX-M β-lactamases are known to hydrolyze cefotaxime more efficiently than ceftazidime, but CTX-M-15 also hydrolyzes ceftazidime efficiently.50 This study showed that there was a significant correlation between expression of the CTX-M-15 gene and resistance to ceftazidime.
The risk factors associated with acquisition of these organisms were not determined in this study that includes: underlying illness, presence of a catheter, prior exposure to beta-lactam and aminoglycosides/fluoroquinolones antibiotic, length of hospitalization, and admission to the intensive care unit.
Conclusion
This study stresses the importance of specific surveillance to optimize empiric treatment of uropathogenic E. coli. Further studies should continue to monitor ESBL-producing bacteria and explore the mechanisms contributing to the spread of CTX-M genotypes in these bacteria to provide reference data to enable relevant infection control.
Disclosure
The authors declared no conflicts of interest. No funding was received for this study.
Acknowledgments
The authors would like to thank Dr. Omar Elahmer and the laboratory staff of the Center for Disease Control for technical support. The authors extend their sincere appreciation to prof. Blackwell for her insightful comments.
references
- 1. Piatti G, Mannini A, Balistreri M, Schito AM. Virulence factors in urinary Escherichia coli strains: phylogenetic background and quinolone and fluoroquinolone resistance. J Clin Microbiol 2008 Feb;46(2):480-487.
- 2. Song S, Lee EY, Koh EM, Ha HS, Jeong HJ, Bae IK, et al. Antibiotic resistance mechanisms of Escherichia coli Isolates from urinary specimens. Korean J Lab Med 2009 Feb;29(1):17-24.
- 3. Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G, et al. CTX-M: changing the face of ESBLs in Europe. J Antimicrob Chemother 2007 Feb;59(2):165-174.
- 4. Doi Y, Park YS, Rivera JI, Adams-Haduch JM, Hingwe A, Sordillo EM, et al. Community-associated extended-spectrum b-lactamase-producing Escherichia coli infection in the United States. Clin Infect Dis 2013 Mar;56(5):641-648.
- 5. Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother 2011 Jan;66(1):1-14.
- 6. Khalaf NG, Eletreby ME, Hanson ND. Characterization of CTX-M ESBLs in Enterobacter cloacae, Escherichia coli and Klebsiella pneumoniae from Cairo, Egypt. BMC Infect Dis 2009;9(84):1-5.
- 7. Ramdani-Bouguessa N, Mendonça N, Leitão J, Ferreira E, Tazir M, Caniça M. CTX-M-3 and CTX-M-15 extended-spectrum b-lactamases in isolates of Escherichia coli from a hospital in Algiers, Algeria. J Clin Microbiol 2006 Dec;44(12):4584-4586.
- 8. Boualiegue-Godet O, Bensalem Y, Fabre L, Demartin M, Grimont PA, Mzoughi R. Nosocomial outbreak caused by Salmonella enteric serotype producing CTX-M-27 extended-spectrum b-lactamase in a neonatal unit in Sousse, Tunisia. J Clin Microbiol 2005;43(3):1843-1849.
- 9. Blomberg B, Jureen R, Manji KP, Tamim BS, Mwakagile DS, Urassa WK, et al. High rate of fatal cases of pediatric septicemia caused by gram-negative bacteria with extended-spectrum b-lactamases in Dar es Salaam, Tanzania. J Clin Microbiol 2005 Feb;43(2):745-749.
- 10. Gangoue-Pieboji J, Miriagou V, Vourli S, Tzelepi E, Ngassam P, Tzouvelekis LS. Emergence of CTX-M-15-producing enterobacteria in Cameroon and characterization of a blaCTX-M-15-carrying element. Antimicrob Agents Chemother 2005 Jan;49(1):441-443.
- 11. Cantón R, González-Alba JM, Galán JC. CTX-M enzymes: origin and diffusion. Front Microbiol 2012 Apr;3:110.
- 12. Poirel L, Bonnin RA, Nordmann P. Genetic support and diversity of acquired extended-spectrum b-lactamases in Gram-negative rods. Infect Genet Evol 2012 Jul;12(5):883-893.
- 13. Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Caniça MM, et al. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 2008 Feb;61(2):273-281.
- 14. Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 2009 Jun;53(6):2227-2238.
- 15. Woodford N, Carattoli A, Karisik E, Underwood A, Ellington MJ, Livermore DM. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob Agents Chemother 2009 Oct;53(10):4472-4482.
- 16. Mnif B, Vimont S, Boyd A, Bourit E, Picard B, Branger C, et al. Molecular characterization of addiction systems of plasmids encoding extended-spectrum beta-lactamases in Escherichia coli. J Antimicrob Chemother 2010 Aug;65(8):1599-1603.
- 17. Doumith M, Dhanji H, Ellington MJ, Hawkey P, Woodford N. Characterization of plasmids encoding extended-spectrum b-lactamases and their addiction systems circulating among Escherichia coli clinical isolates in the UK. J Antimicrob Chemother 2012 Apr;67(4):878-885.
- 18. Gerdes K, Christensen SK, Løbner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol 2005 May;3(5):371-382.
- 19. Abujnah AA, Zorgani A, Sabri MA, El-Mohammady H, Khalek RA, Ghenghesh KS. Multidrug resistance and extended-spectrum b-lactamases genes among Escherichia coli from patients with urinary tract infections in Northwestern Libya. Libyan J Med 2015 Feb;10:26412.
- 20. Kraiem AG. Zorgani A, Elahmer O, Hammami A, Shaaben B, Ghenghesh KS. New Delhi metallo-b-lactamase (NDM) and OXA-48 carbapenemases in Gram-negative bacilli isolates in Libya. Libyan J Med 2015 Aug;10(1):29206.
- 21. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-second Informational Supplement M100-S22. CLSI, Wayne, PA, USA, 2012.
- 22. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012 Mar;18(3):268-281.
- 23. Leflon-Guibout V, Jurand C, Bonacorsi S, Espinasse F, Guelfi MC, Duportail F, et al. Emergence and spread of three clonally related virulent isolates of CTX-M-15-producing Escherichia coli with variable resistance to aminoglycosides and tetracycline in a French geriatric hospital. Antimicrob Agents Chemother 2004 Oct;48(10):3736-3742.
- 24. Al-Otaibi FE, Bukhari EE. Clinical and laboratory profiles of urinary tract infections caused by extended-spectrum beta-lactamase-producing Escherichia coli in a tertiary care center in central Saudi Arabia. Saudi Med J 2013 Feb;34(2):171-176.
- 25. Ejikeugwu PC, Ikegbunam NM, Ugwu CM, Iroha IR, Esimone CO. Extended-spectrum β-lactamase-producing Escherichia coli isolates from suspected community acquired urinary tract infections. Eur J Sci Res 2012;84(2):565-571.
- 26. Hawkey PM. Prevalence and clonality of extended-spectrum beta-lactamases in Asia. Clin Microbiol Infect 2008 Jan;14(Suppl 1):159-165.
- 27. Perez F, Endimiani A, Hujer KM, Bonomo RA. The continuing challenge of ESBLs. Curr Opin Pharmacol 2007 Oct;7(5):459-469.
- 28. Feizabadi MM, Delfani S, Raji N, Majnooni A, Aligholi M, Shahcheraghi F, et al. Distribution of bla(TEM), bla(SHV), bla(CTX-M) genes among clinical isolates of Klebsiella pneumoniae at Labbafinejad Hospital, Tehran, Iran. Microb Drug Resist 2010 Mar;16(1):49-53.
- 29. Morrissey I, Hackel M, Badal R, Bouchillon S, Hawser S, Biedenbach D. A review of ten years of the study for monitoring antimicrobial resistance trends (SMART) from 2002 to 2011. Pharmaceuticals (Basel) 2013 Nov;6(11):1335-1346.
- 30. Daoud Z, Afif C. Escherichia coli isolated from urinary tract infections of Lebanese patients between 2000 and 2009: epidemiology and profiles of resistance. Chemother Res Pract 2011;2011:218431.
- 31. Al-Agamy MH, Shibl AM, Hafez MM, Al-Ahdal MN, Memish ZA, Khubnani H. Molecular characteristics of extended-spectrum b-lactamase-producing Escherichia coli in Riyadh: emergence of CTX-M-15-producing E. coli ST131. Ann Clin Microbiol Antimicrob 2014 Jan;13:4.
- 32. Hassan H, Abdalhamid B. Molecular characterization of extended-spectrum beta-lactamase producing Enterobacteriaceae in a Saudi Arabian tertiary hospital. J Infect Dev Ctries 2014 Mar;8(3):282-288.
- 33. Coque TM, Baquero F, Canton R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill 2008 Nov;13(47):19044.
- 34. Pitout JD, Laupland KB. Extended-spectrum b-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 2008 Mar;8(3):159-166.
- 35. Brigante G, Luzzaro F, Perilli M, Lombardi G, Colì A, Rossolini GM, et al. Evolution of CTX-M-type b-lactamases in isolates of Escherichia coli infecting hospital and community patients. Int J Antimicrob Agents 2005 Feb;25(2):157-162.
- 36. Cantón R, Coque TM. The CTX-M b-lactamase pandemic. Curr Opin Microbiol 2006 Oct;9(5):466-475.
- 37. Sonnevend A, Al Dhaheri K, Mag T, Herpay M, Kolodziejek J, Nowotny N, et al. CTX-M-15-producing multidrug-resistant enteroaggregative Escherichia coli in the United Arab Emirates. Clin Microbiol Infect 2006 Jun;12(6):582-585.
- 38. Ensor VM, Jamal W, Rotimi VO, Evans JT, Hawkey PM. Predominance of CTX-M-15 extended spectrum beta-lactamases in diverse Escherichia coli and Klebsiella pneumoniae from hospital and community patients in Kuwait. Int J Antimicrob Agents 2009 May;33(5):487-489.
- 39. Mohamed Al-Agamy MH, El-Din Ashour MS, Wiegand I. First description of CTX-M beta-lactamase-producing clinical Escherichia coli isolates from Egypt. Int J Antimicrob Agents 2006 Jun;27(6):545-548.
- 40. Mamlouk K, Boutiba-Ben Boubaker I, Gautier V, Vimont S, Picard B, Ben Redjeb S, et al. Emergence and outbreaks of CTX-M beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae strains in a Tunisian hospital. J Clin Microbiol 2006 Nov;44(11):4049-4056.
- 41. Ben Slama K, Ben Sallem R, Jouini A, Rachid S, Moussa L, Sáenz Y, et al. Diversity of genetic lineages among CTX-M-15 and CTX-M-14 producing Escherichia coli strains in a Tunisian hospital. Curr Microbiol 2011 Jun;62(6):1794-1801.
- 42. Mnif B, Harhour H, Jdidi J, Mahjoubi F, Genel N, Arlet G, et al. Molecular epidemiology of extended-spectrum beta-lactamase-producing Escherichia coli in Tunisia and characterization of their virulence factors and plasmid addiction systems. BMC Microbiol 2013 Jun;13 (1):147.
- 43. Zamani K, Emami A, Bazargani A, Moattari A. Phenotypic and molecular characterization of CTX-M extended-spectrum beta-lactamase-producing Escherichia coli isolates in Shiraz, Iran. Rev Soc Bras Med Trop 2015 Jul-Aug;48(4):479-482.
- 44. Hayajneh WA, Hajj A, Hulliel F, Sarkis DK, Irani-Hakimeh N, Kazan L, et al. Susceptibility trends and molecular characterization of Gram-negative bacilli associated with urinary tract and intra-abdominal infections in Jordan and Lebanon: SMART 2011-2013. Int J Infect Dis 2015 Jun;35:56-61.
- 45. Moubareck C, Daoud Z, Hakimé NI, Hamzé M, Mangeney N, Matta H, et al. Countrywide spread of community- and hospital-acquired extended-spectrum beta-lactamase (CTX-M-15)-producing Enterobacteriaceae in Lebanon. J Clin Microbiol 2005 Jul;43(7):3309-3313.
- 46. Xia S, Fan X, Huang Z, Xia L, Xiao M, Chen R, et al. Dominance of CTX-M-type extended-spectrum b-lactamase (ESBL)-producing Escherichia coli isolated from patients with community-onset and hospital-onset infection in China. PLoS One 2014 Jul;9(7):e100707.
- 47. Sennati S, Santella G, Di Conza J, Pallecchi L, Pino M, Ghiglione B, et al. Changing epidemiology of extended-spectrum b-lactamases in Argentina: emergence of CTX-M-15. Antimicrob Agents Chemother 2012 Nov;56(11):6003-6005.
- 48. Giedraitienė A, Vitkauskienė A, Ašmonienė V, Plančiūnienė R, Simonytė S, Pavilonis A, et al. CTX-M-producing Escherichia coli in Lithuania: associations between sites of infection, coresistance, and phylogenetic groups. Medicina (Kaunas) 2013;49(9):393-398.
- 49. Alzohairy M, Khadri H. Frequency and antibiotic susceptibility pattern of uropathogens isolated from community and hospital acquired infections in Saudi Arabia-a prospective case study. Brit J Med Res 2011 Apr-Jun;1(2):45-56.
- 50. Poirel L, Gniadkowski M, Nordmann P. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum b-lactamase CTX-M-15 and of its structurally related b-lactamase CTX-M-3. J Antimicrob Chemother 2002 Dec;50(6):1031-1034.